Jingxing Wu
VDDB: a comprehensive resource and machine learning platform for antiviral drug discovery
Sep 17, 2022



Abstract:Virus infection is one of the major diseases that seriously threaten human health. To meet the growing demand for mining and sharing data resources related to antiviral drugs and to accelerate the design and discovery of new antiviral drugs, we presented an open-access antiviral drug resource and machine learning platform (VDDB), which, to the best of our knowledge, is the first comprehensive dedicated resource for experimentally verified potential drugs/molecules based on manually curated data. Currently, VDDB highlights 848 clinical vaccines, 199 clinical antibodies, as well as over 710,000 small molecules targeting 39 medically important viruses including SARS-CoV-2. Furthermore, VDDB stores approximately 3 million records of pharmacological data for these collected potential antiviral drugs/molecules, involving 314 cell infection-based phenotypic and 234 target-based genotypic assays. Based on these annotated pharmacological data, VDDB allows users to browse, search and download reliable information about these collects for various viruses of interest. In particular, VDDB also integrates 57 cell infection- and 117 target-based associated high-accuracy machine learning models to support various antivirals identification-related tasks, such as compound activity prediction, virtual screening, drug repositioning and target fishing. VDDB is freely accessible at http://vddb.idruglab.cn.
FP-GNN: a versatile deep learning architecture for enhanced molecular property prediction
May 08, 2022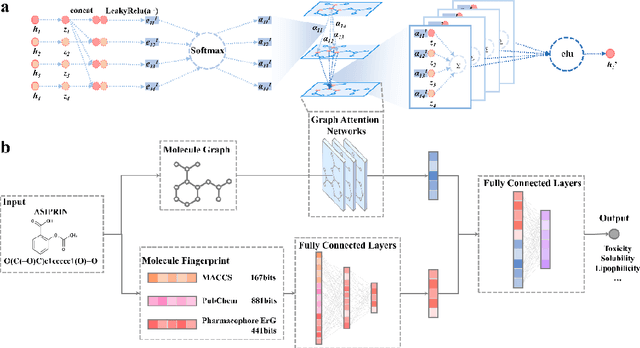
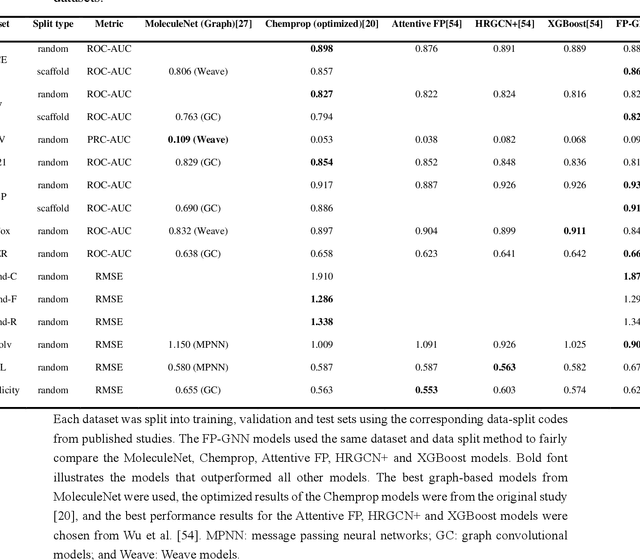
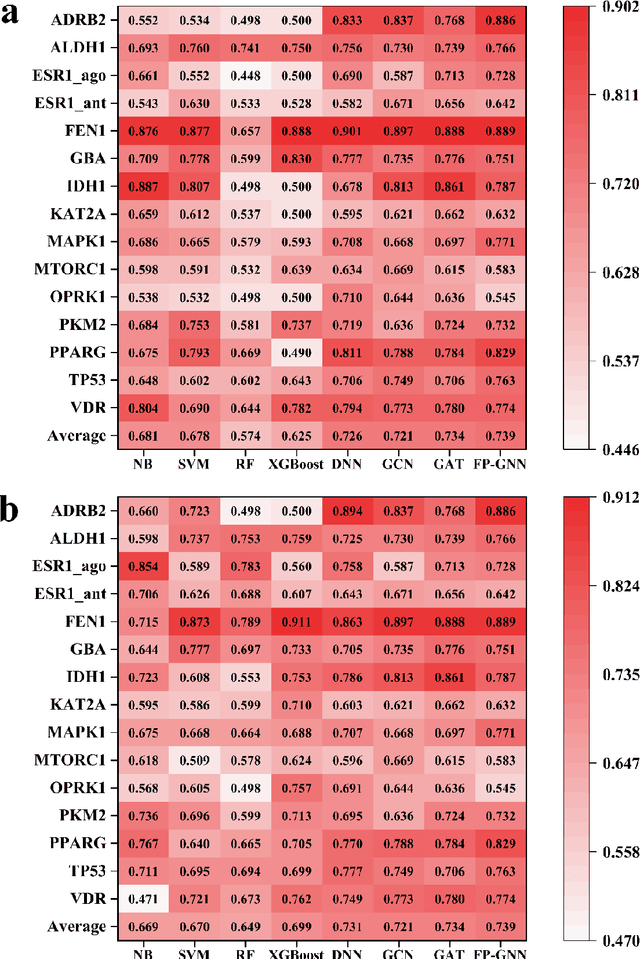
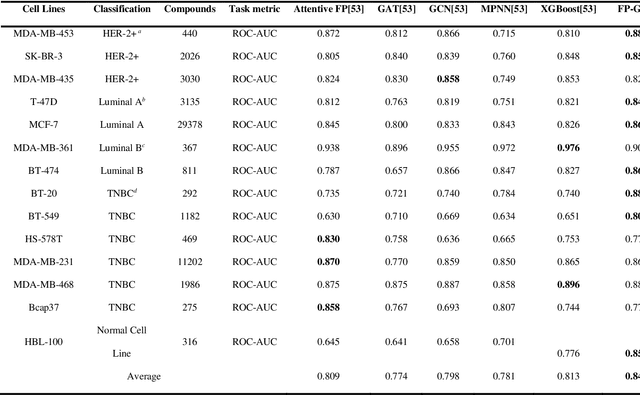
Abstract:Deep learning is an important method for molecular design and exhibits considerable ability to predict molecular properties, including physicochemical, bioactive, and ADME/T (absorption, distribution, metabolism, excretion, and toxicity) properties. In this study, we advanced a novel deep learning architecture, termed FP-GNN, which combined and simultaneously learned information from molecular graphs and fingerprints. To evaluate the FP-GNN model, we conducted experiments on 13 public datasets, an unbiased LIT-PCBA dataset, and 14 phenotypic screening datasets for breast cell lines. Extensive evaluation results showed that compared to advanced deep learning and conventional machine learning algorithms, the FP-GNN algorithm achieved state-of-the-art performance on these datasets. In addition, we analyzed the influence of different molecular fingerprints, and the effects of molecular graphs and molecular fingerprints on the performance of the FP-GNN model. Analysis of the anti-noise ability and interpretation ability also indicated that FP-GNN was competitive in real-world situations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge