Jan Hasenauer
Life and Medical Sciences
Diffusion Models in Simulation-Based Inference: A Tutorial Review
Dec 22, 2025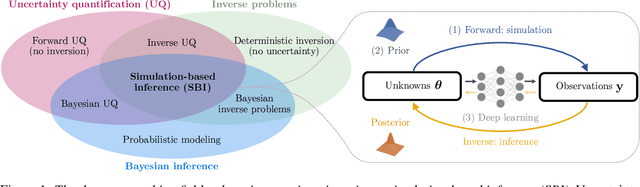
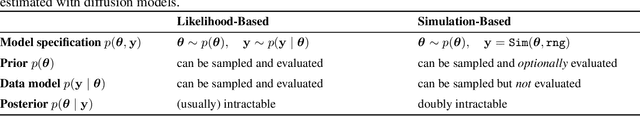
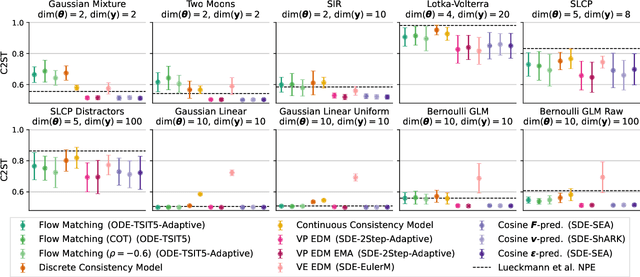
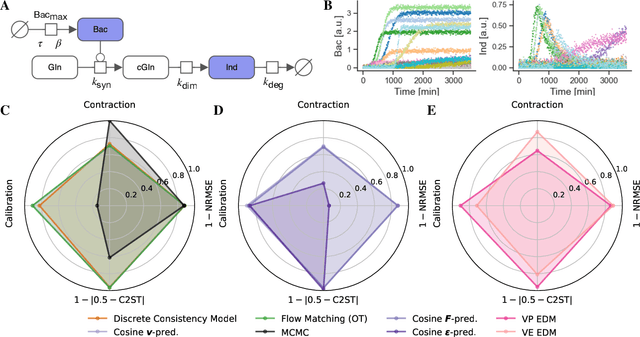
Abstract:Diffusion models have recently emerged as powerful learners for simulation-based inference (SBI), enabling fast and accurate estimation of latent parameters from simulated and real data. Their score-based formulation offers a flexible way to learn conditional or joint distributions over parameters and observations, thereby providing a versatile solution to various modeling problems. In this tutorial review, we synthesize recent developments on diffusion models for SBI, covering design choices for training, inference, and evaluation. We highlight opportunities created by various concepts such as guidance, score composition, flow matching, consistency models, and joint modeling. Furthermore, we discuss how efficiency and statistical accuracy are affected by noise schedules, parameterizations, and samplers. Finally, we illustrate these concepts with case studies across parameter dimensionalities, simulation budgets, and model types, and outline open questions for future research.
Scalable branch-and-bound model selection with non-monotonic criteria including AIC, BIC and Mallows's $\mathit{C_p}$
Dec 13, 2025Abstract:Model selection is a pivotal process in the quantitative sciences, where researchers must navigate between numerous candidate models of varying complexity. Traditional information criteria, such as the corrected Akaike Information Criterion (AICc), Bayesian Information Criterion (BIC), and Mallows's $\mathit{C_p}$, are valuable tools for identifying optimal models. However, the exponential increase in candidate models with each additional model parameter renders the evaluation of these criteria for all models -- a strategy known as exhaustive, or brute-force, searches -- computationally prohibitive. Consequently, heuristic approaches like stepwise regression are commonly employed, albeit without guarantees of finding the globally-optimal model. In this study, we challenge the prevailing notion that non-monotonicity in information criteria precludes bounds on the search space. We introduce a simple but novel bound that enables the development of branch-and-bound algorithms tailored for these non-monotonic functions. We demonstrate that our approach guarantees identification of the optimal model(s) across diverse model classes, sizes, and applications, often with orders of magnitude computational speedups. For instance, in one previously-published model selection task involving $2^{32}$ (approximately 4 billion) candidate models, our method achieves a computational speedup exceeding 6,000. These findings have broad implications for the scalability and effectiveness of model selection in complex scientific domains.
Non-Negative Universal Differential Equations With Applications in Systems Biology
Jun 20, 2024Abstract:Universal differential equations (UDEs) leverage the respective advantages of mechanistic models and artificial neural networks and combine them into one dynamic model. However, these hybrid models can suffer from unrealistic solutions, such as negative values for biochemical quantities. We present non-negative UDE (nUDEs), a constrained UDE variant that guarantees non-negative values. Furthermore, we explore regularisation techniques to improve generalisation and interpretability of UDEs.
Assessment of Uncertainty Quantification in Universal Differential Equations
Jun 13, 2024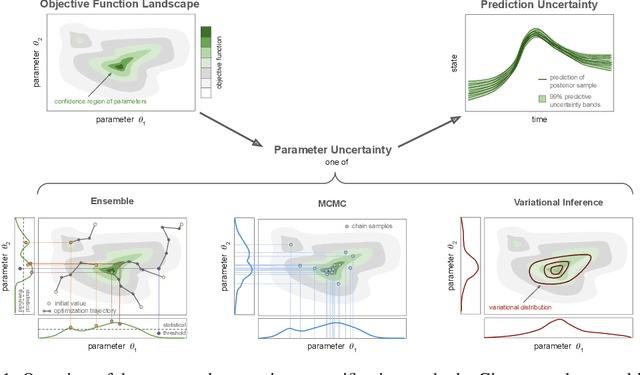
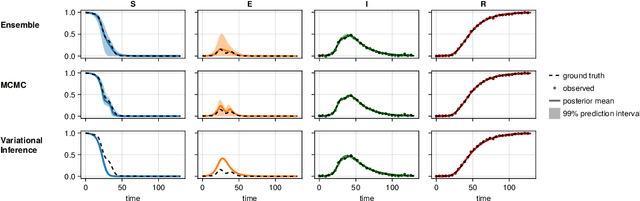
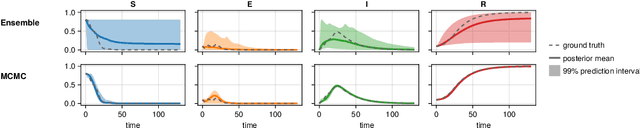
Abstract:Scientific Machine Learning is a new class of approaches that integrate physical knowledge and mechanistic models with data-driven techniques for uncovering governing equations of complex processes. Among the available approaches, Universal Differential Equations (UDEs) are used to combine prior knowledge in the form of mechanistic formulations with universal function approximators, like neural networks. Integral to the efficacy of UDEs is the joint estimation of parameters within mechanistic formulations and the universal function approximators using empirical data. The robustness and applicability of resultant models, however, hinge upon the rigorous quantification of uncertainties associated with these parameters, as well as the predictive capabilities of the overall model or its constituent components. With this work, we provide a formalisation of uncertainty quantification (UQ) for UDEs and investigate important frequentist and Bayesian methods. By analysing three synthetic examples of varying complexity, we evaluate the validity and efficiency of ensembles, variational inference and Markov chain Monte Carlo sampling as epistemic UQ methods for UDEs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge