Jan Franz Nygård
A Multiparty Homomorphic Encryption Approach to Confidential Federated Kaplan Meier Survival Analysis
Dec 29, 2024Abstract:The proliferation of healthcare data has expanded opportunities for collaborative research, yet stringent privacy regulations hinder pooling sensitive patient records. We propose a \emph{multiparty homomorphic encryption-based} framework for \emph{privacy-preserving federated Kaplan--Meier survival analysis}, offering native floating-point support, a theoretical model, and explicit reconstruction-attack mitigation. Compared to prior work, our framework ensures encrypted federated survival estimates closely match centralized outcomes, supported by formal utility-loss bounds that demonstrate convergence as aggregation and decryption noise diminish. Extensive experiments on the NCCTG Lung Cancer and synthetic Breast Cancer datasets confirm low \emph{mean absolute error (MAE)} and \emph{root mean squared error (RMSE)}, indicating negligible deviations between encrypted and non-encrypted survival curves. Log-rank and numerical accuracy tests reveal \emph{no significant difference} between federated encrypted and non-encrypted analyses, preserving statistical validity. A reconstruction-attack evaluation shows smaller federations (2--3 providers) with overlapping data between the institutions are vulnerable, a challenge mitigated by multiparty encryption. Larger federations (5--50 sites) degrade reconstruction accuracy further, with encryption improving confidentiality. Despite an 8--19$\times$ computational overhead, threshold-based homomorphic encryption is \emph{feasible for moderate-scale deployments}, balancing security and runtime. By providing robust privacy guarantees alongside high-fidelity survival estimates, our framework advances the state-of-the art in secure multi-institutional survival analysis.
A Differentially Private Kaplan-Meier Estimator for Privacy-Preserving Survival Analysis
Dec 06, 2024
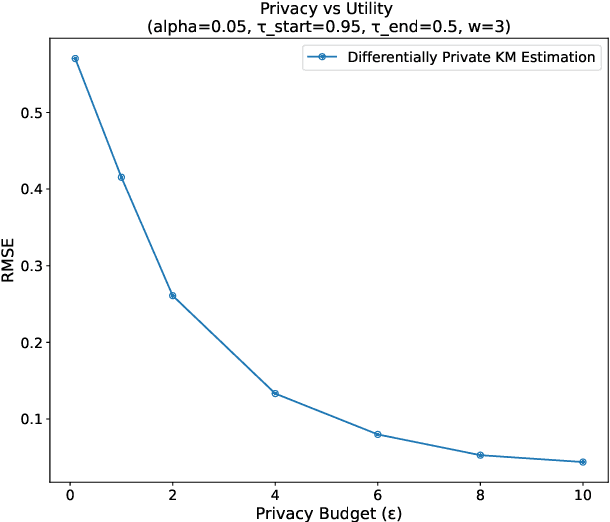
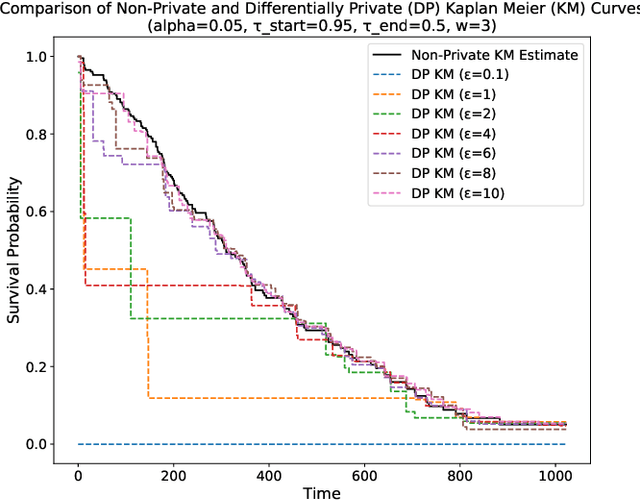
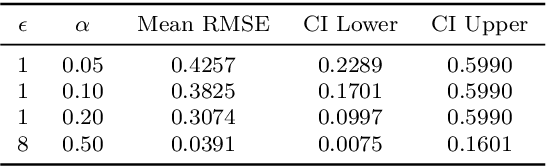
Abstract:This paper presents a differentially private approach to Kaplan-Meier estimation that achieves accurate survival probability estimates while safeguarding individual privacy. The Kaplan-Meier estimator is widely used in survival analysis to estimate survival functions over time, yet applying it to sensitive datasets, such as clinical records, risks revealing private information. To address this, we introduce a novel algorithm that applies time-indexed Laplace noise, dynamic clipping, and smoothing to produce a privacy-preserving survival curve while maintaining the cumulative structure of the Kaplan-Meier estimator. By scaling noise over time, the algorithm accounts for decreasing sensitivity as fewer individuals remain at risk, while dynamic clipping and smoothing prevent extreme values and reduce fluctuations, preserving the natural shape of the survival curve. Our results, evaluated on the NCCTG lung cancer dataset, show that the proposed method effectively lowers root mean squared error (RMSE) and enhances accuracy across privacy budgets ($\epsilon$). At $\epsilon = 10$, the algorithm achieves an RMSE as low as 0.04, closely approximating non-private estimates. Additionally, membership inference attacks reveal that higher $\epsilon$ values (e.g., $\epsilon \geq 6$) significantly reduce influential points, particularly at higher thresholds, lowering susceptibility to inference attacks. These findings confirm that our approach balances privacy and utility, advancing privacy-preserving survival analysis.
Can I trust my fake data -- A comprehensive quality assessment framework for synthetic tabular data in healthcare
Jan 24, 2024



Abstract:Ensuring safe adoption of AI tools in healthcare hinges on access to sufficient data for training, testing and validation. In response to privacy concerns and regulatory requirements, using synthetic data has been suggested. Synthetic data is created by training a generator on real data to produce a dataset with similar statistical properties. Competing metrics with differing taxonomies for quality evaluation have been suggested, resulting in a complex landscape. Optimising quality entails balancing considerations that make the data fit for use, yet relevant dimensions are left out of existing frameworks. We performed a comprehensive literature review on the use of quality evaluation metrics on SD within the scope of tabular healthcare data and SD made using deep generative methods. Based on this and the collective team experiences, we developed a conceptual framework for quality assurance. The applicability was benchmarked against a practical case from the Dutch National Cancer Registry. We present a conceptual framework for quality assurance of SD for AI applications in healthcare that aligns diverging taxonomies, expands on common quality dimensions to include the dimensions of Fairness and Carbon footprint, and proposes stages necessary to support real-life applications. Building trust in synthetic data by increasing transparency and reducing the safety risk will accelerate the development and uptake of trustworthy AI tools for the benefit of patients. Despite the growing emphasis on algorithmic fairness and carbon footprint, these metrics were scarce in the literature review. The overwhelming focus was on statistical similarity using distance metrics while sequential logic detection was scarce. A consensus-backed framework that includes all relevant quality dimensions can provide assurance for safe and responsible real-life applications of SD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge