Hsing-Kuo Pao
Dept. of Computer Science and Information Engineering National Taiwan University of Science and Technology, Taipei, Taiwan
Parallel Swin Transformer-Enhanced 3D MRI-to-CT Synthesis for MRI-Only Radiotherapy Planning
Feb 05, 2026Abstract:MRI provides superior soft tissue contrast without ionizing radiation; however, the absence of electron density information limits its direct use for dose calculation. As a result, current radiotherapy workflows rely on combined MRI and CT acquisitions, increasing registration uncertainty and procedural complexity. Synthetic CT generation enables MRI only planning but remains challenging due to nonlinear MRI-CT relationships and anatomical variability. We propose Parallel Swin Transformer-Enhanced Med2Transformer, a 3D architecture that integrates convolutional encoding with dual Swin Transformer branches to model both local anatomical detail and long-range contextual dependencies. Multi-scale shifted window attention with hierarchical feature aggregation improves anatomical fidelity. Experiments on public and clinical datasets demonstrate higher image similarity and improved geometric accuracy compared with baseline methods. Dosimetric evaluation shows clinically acceptable performance, with a mean target dose error of 1.69%. Code is available at: https://github.com/mobaidoctor/med2transformer.
Data Complexity-aware Deep Model Performance Forecasting
Jan 04, 2026Abstract:Deep learning models are widely used across computer vision and other domains. When working on the model induction, selecting the right architecture for a given dataset often relies on repetitive trial-and-error procedures. This procedure is time-consuming, resource-intensive, and difficult to automate. While previous work has explored performance prediction using partial training or complex simulations, these methods often require significant computational overhead or lack generalizability. In this work, we propose an alternative approach: a lightweight, two-stage framework that can estimate model performance before training given the understanding of the dataset and the focused deep model structures. The first stage predicts a baseline based on the analysis of some measurable properties of the dataset, while the second stage adjusts the estimation with additional information on the model's architectural and hyperparameter details. The setup allows the framework to generalize across datasets and model types. Moreover, we find that some of the underlying features used for prediction - such as dataset variance - can offer practical guidance for model selection, and can serve as early indicators of data quality. As a result, the framework can be used not only to forecast model performance, but also to guide architecture choices, inform necessary preprocessing procedures, and detect potentially problematic datasets before training begins.
Large Language Model Pruning
May 24, 2024Abstract:We surely enjoy the larger the better models for their superior performance in the last couple of years when both the hardware and software support the birth of such extremely huge models. The applied fields include text mining and others. In particular, the success of LLMs on text understanding and text generation draws attention from researchers who have worked on NLP and related areas for years or even decades. On the side, LLMs may suffer from problems like model overfitting, hallucination, and device limitation to name a few. In this work, we suggest a model pruning technique specifically focused on LLMs. The proposed methodology emphasizes the explainability of deep learning models. By having the theoretical foundation, we obtain a trustworthy deep model so that huge models with a massive number of model parameters become not quite necessary. A mutual information-based estimation is adopted to find neurons with redundancy to eliminate. Moreover, an estimator with well-tuned parameters helps to find precise estimation to guide the pruning procedure. At the same time, we also explore the difference between pruning on large-scale models vs. pruning on small-scale models. The choice of pruning criteria is sensitive in small models but not for large-scale models. It is a novel finding through this work. Overall, we demonstrate the superiority of the proposed model to the state-of-the-art models.
Polyp-DDPM: Diffusion-Based Semantic Polyp Synthesis for Enhanced Segmentation
Feb 06, 2024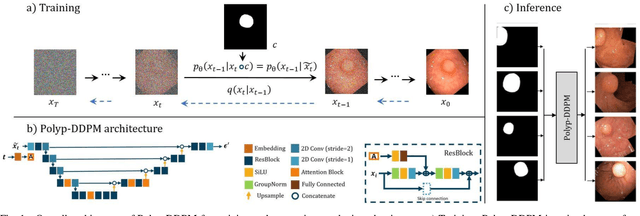
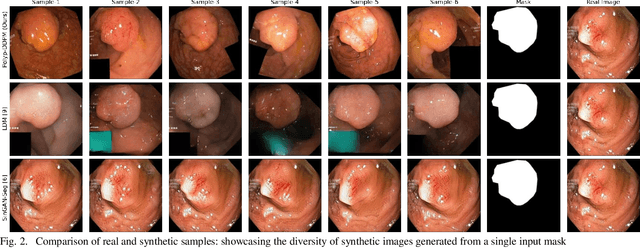
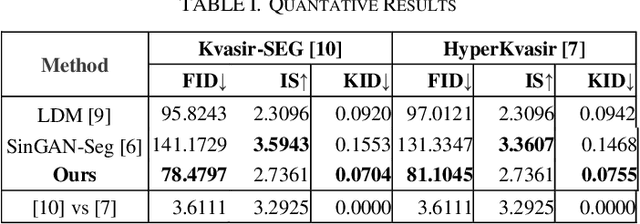
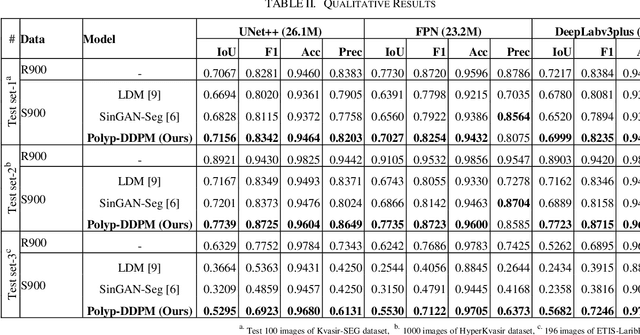
Abstract:This study introduces Polyp-DDPM, a diffusion-based method for generating realistic images of polyps conditioned on masks, aimed at enhancing the segmentation of gastrointestinal (GI) tract polyps. Our approach addresses the challenges of data limitations, high annotation costs, and privacy concerns associated with medical images. By conditioning the diffusion model on segmentation masks-binary masks that represent abnormal areas-Polyp-DDPM outperforms state-of-the-art methods in terms of image quality (achieving a Frechet Inception Distance (FID) score of 78.47, compared to scores above 83.79) and segmentation performance (achieving an Intersection over Union (IoU) of 0.7156, versus less than 0.6694 for synthetic images from baseline models and 0.7067 for real data). Our method generates a high-quality, diverse synthetic dataset for training, thereby enhancing polyp segmentation models to be comparable with real images and offering greater data augmentation capabilities to improve segmentation models. The source code and pretrained weights for Polyp-DDPM are made publicly available at https://github.com/mobaidoctor/polyp-ddpm.
Conditional Diffusion Models for Semantic 3D Medical Image Synthesis
May 29, 2023



Abstract:This paper introduces Med-DDPM, an innovative solution using diffusion models for semantic 3D medical image synthesis, addressing the prevalent issues in medical imaging such as data scarcity, inconsistent acquisition methods, and privacy concerns. Experimental evidence illustrates that diffusion models surpass Generative Adversarial Networks (GANs) in stability and performance, generating high-quality, realistic 3D medical images. The distinct feature of Med-DDPM is its use of semantic conditioning for the diffusion model in 3D image synthesis. By controlling the generation process through pixel-level mask labels, it facilitates the creation of realistic medical images. Empirical evaluations underscore the superior performance of Med-DDPM over GAN techniques in metrics such as accuracy, stability, and versatility. Furthermore, Med-DDPM outperforms traditional augmentation techniques and synthetic GAN images in enhancing the accuracy of segmentation models. It addresses challenges such as insufficient datasets, lack of annotated data, and class imbalance. Noting the limitations of the Frechet inception distance (FID) metric, we introduce a histogram-equalized FID metric for effective performance evaluation. In summary, Med-DDPM, by utilizing diffusion models, signifies a crucial step forward in the domain of high-resolution semantic 3D medical image synthesis, transcending the limitations of GANs and data constraints. This method paves the way for a promising solution in medical imaging, primarily for data augmentation and anonymization, thus contributing significantly to the field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge