Hongyaoxing Gu
LoPRo: Enhancing Low-Rank Quantization via Permuted Block-Wise Rotation
Jan 27, 2026Abstract:Post-training quantization (PTQ) enables effective model compression while preserving relatively high accuracy. Current weight-only PTQ methods primarily focus on the challenging sub-3-bit regime, where approaches often suffer significant accuracy degradation, typically requiring fine-tuning to achieve competitive performance. In this work, we revisit the fundamental characteristics of weight quantization and analyze the challenges in quantizing the residual matrix under low-rank approximation. We propose LoPRo, a novel fine-tuning-free PTQ algorithm that enhances residual matrix quantization by applying block-wise permutation and Walsh-Hadamard transformations to rotate columns of similar importance, while explicitly preserving the quantization accuracy of the most salient column blocks. Furthermore, we introduce a mixed-precision fast low-rank decomposition based on rank-1 sketch (R1SVD) to further minimize quantization costs. Experiments demonstrate that LoPRo outperforms existing fine-tuning-free PTQ methods at both 2-bit and 3-bit quantization, achieving accuracy comparable to fine-tuning baselines. Specifically, LoPRo achieves state-of-the-art quantization accuracy on LLaMA-2 and LLaMA-3 series models while delivering up to a 4$\times$ speedup. In the MoE model Mixtral-8x7B, LoPRo completes quantization within 2.5 hours, simultaneously reducing perplexity by 0.4$\downarrow$ and improving accuracy by 8\%$\uparrow$. Moreover, compared to other low-rank quantization methods, LoPRo achieves superior accuracy with a significantly lower rank, while maintaining high inference efficiency and minimal additional latency.
A method of using RSVD in residual calculation of LowBit GEMM
Sep 27, 2024Abstract:The advancements of hardware technology in recent years has brought many possibilities for low-precision applications. However, the use of low precision can introduce significant computational errors, posing a considerable challenge to maintaining the computational accuracy. We propose low-rank residuals quantized matrix multiplication(LRQMM) method which introduces low-rank approximation in residual compensation for dense low precision quantization matrix multiplication. It can bring several times accuracy improvement with only BLAS-2 level extra time overhead. Moreover, LRQMM is a completely data-free quantization method that does not require additional data for pre-training. And it only works with low precision GEMM operator, which is easy to couple with other methods. Through experimentation, LRQMM can reduce the error of direct quantized matrix multiplication by 1~2 orders of magnitude, when dealing with larger matrix sizes, the computational speed is only reduced by approximately 20\%. In deep learning networks, LRQMM-4bit achieves 61.8% ImageNet Top-1 accuracy in Resnet-50, while the Direct Quant accuracy is only 8.3%.
A differentiable brain simulator bridging brain simulation and brain-inspired computing
Nov 09, 2023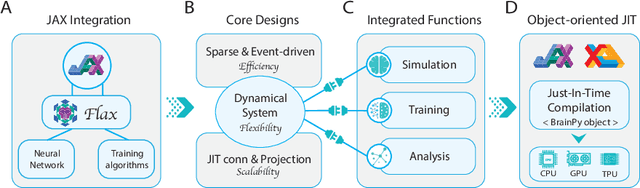

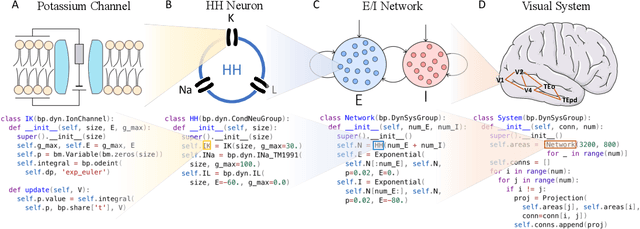

Abstract:Brain simulation builds dynamical models to mimic the structure and functions of the brain, while brain-inspired computing (BIC) develops intelligent systems by learning from the structure and functions of the brain. The two fields are intertwined and should share a common programming framework to facilitate each other's development. However, none of the existing software in the fields can achieve this goal, because traditional brain simulators lack differentiability for training, while existing deep learning (DL) frameworks fail to capture the biophysical realism and complexity of brain dynamics. In this paper, we introduce BrainPy, a differentiable brain simulator developed using JAX and XLA, with the aim of bridging the gap between brain simulation and BIC. BrainPy expands upon the functionalities of JAX, a powerful AI framework, by introducing complete capabilities for flexible, efficient, and scalable brain simulation. It offers a range of sparse and event-driven operators for efficient and scalable brain simulation, an abstraction for managing the intricacies of synaptic computations, a modular and flexible interface for constructing multi-scale brain models, and an object-oriented just-in-time compilation approach to handle the memory-intensive nature of brain dynamics. We showcase the efficiency and scalability of BrainPy on benchmark tasks, highlight its differentiable simulation for biologically plausible spiking models, and discuss its potential to support research at the intersection of brain simulation and BIC.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge