Henri Palacci
Partial Product Aware Machine Learning on DNA-Encoded Libraries
May 16, 2022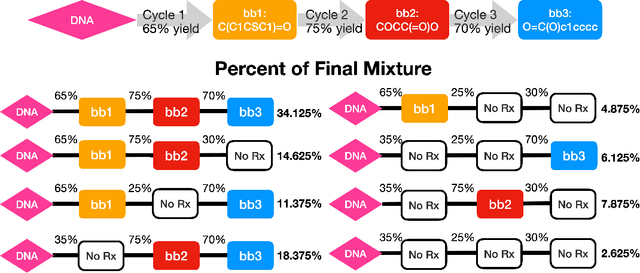
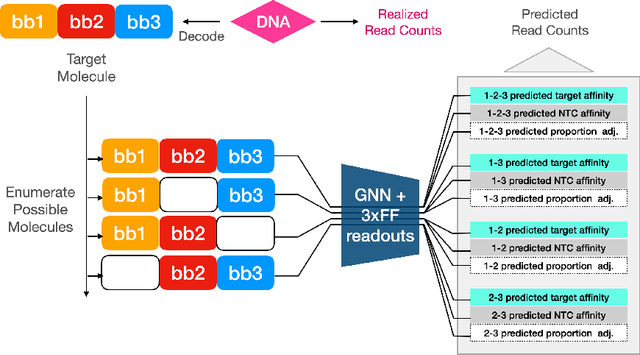
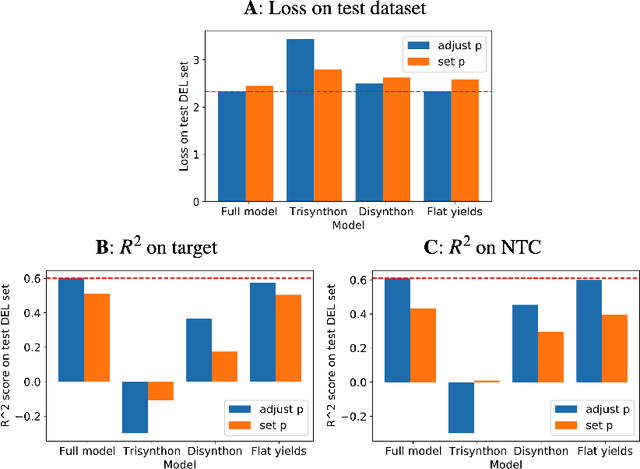
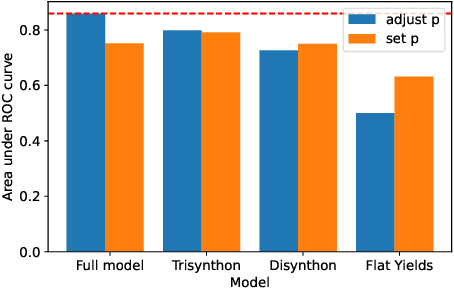
Abstract:DNA encoded libraries (DELs) are used for rapid large-scale screening of small molecules against a protein target. These combinatorial libraries are built through several cycles of chemistry and DNA ligation, producing large sets of DNA-tagged molecules. Training machine learning models on DEL data has been shown to be effective at predicting molecules of interest dissimilar from those in the original DEL. Machine learning chemical property prediction approaches rely on the assumption that the property of interest is linked to a single chemical structure. In the context of DNA-encoded libraries, this is equivalent to assuming that every chemical reaction fully yields the desired product. However, in practice, multi-step chemical synthesis sometimes generates partial molecules. Each unique DNA tag in a DEL therefore corresponds to a set of possible molecules. Here, we leverage reaction yield data to enumerate the set of possible molecules corresponding to a given DNA tag. This paper demonstrates that training a custom GNN on this richer dataset improves accuracy and generalization performance.
Scalable Natural Gradient Langevin Dynamics in Practice
Jun 07, 2018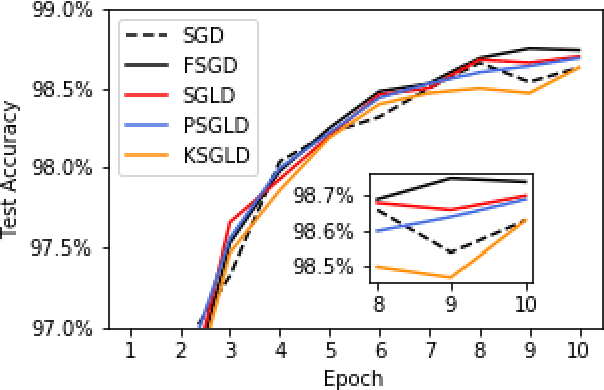
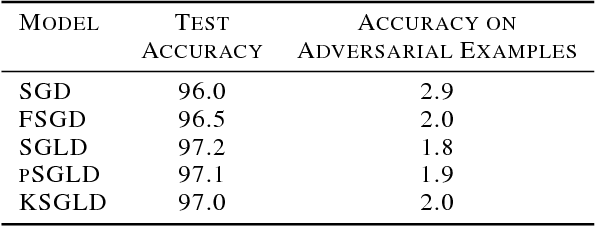
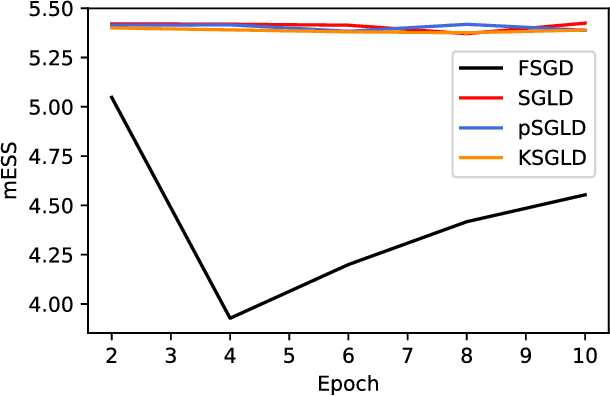
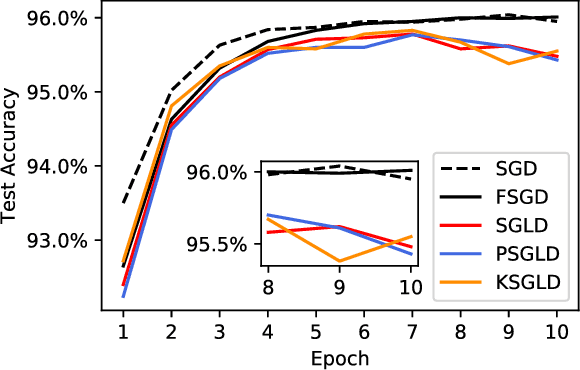
Abstract:Stochastic Gradient Langevin Dynamics (SGLD) is a sampling scheme for Bayesian modeling adapted to large datasets and models. SGLD relies on the injection of Gaussian Noise at each step of a Stochastic Gradient Descent (SGD) update. In this scheme, every component in the noise vector is independent and has the same scale, whereas the parameters we seek to estimate exhibit strong variations in scale and significant correlation structures, leading to poor convergence and mixing times. We compare different preconditioning approaches to the normalization of the noise vector and benchmark these approaches on the following criteria: 1) mixing times of the multivariate parameter vector, 2) regularizing effect on small dataset where it is easy to overfit, 3) covariate shift detection and 4) resistance to adversarial examples.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge