Heinrich Martin Overhoff
PCA for Enhanced Cross-Dataset Generalizability in Breast Ultrasound Tumor Segmentation
May 29, 2025Abstract:In medical image segmentation, limited external validity remains a critical obstacle when models are deployed across unseen datasets, an issue particularly pronounced in the ultrasound image domain. Existing solutions-such as domain adaptation and GAN-based style transfer-while promising, often fall short in the medical domain where datasets are typically small and diverse. This paper presents a novel application of principal component analysis (PCA) to address this limitation. PCA preprocessing reduces noise and emphasizes essential features by retaining approximately 90\% of the dataset variance. We evaluate our approach across six diverse breast tumor ultrasound datasets comprising 3,983 B-mode images and corresponding expert tumor segmentation masks. For each dataset, a corresponding dimensionality reduced PCA-dataset is created and U-Net-based segmentation models are trained on each of the twelve datasets. Each model trained on an original dataset was inferenced on the remaining five out-of-domain original datasets (baseline results), while each model trained on a PCA dataset was inferenced on five out-of-domain PCA datasets. Our experimental results indicate that using PCA reconstructed datasets, instead of original images, improves the model's recall and Dice scores, particularly for model-dataset pairs where baseline performance was lowest, achieving statistically significant gains in recall (0.57 $\pm$ 0.07 vs. 0.70 $\pm$ 0.05, $p = 0.0004$) and Dice scores (0.50 $\pm$ 0.06 vs. 0.58 $\pm$ 0.06, $p = 0.03$). Our method reduced the decline in recall values due to external validation by $33\%$. These findings underscore the potential of PCA reconstruction as a safeguard to mitigate declines in segmentation performance, especially in challenging cases, with implications for enhancing external validity in real-world medical applications.
Impact of PCA-based preprocessing and different CNN structures on deformable registration of sonograms
Jan 20, 2023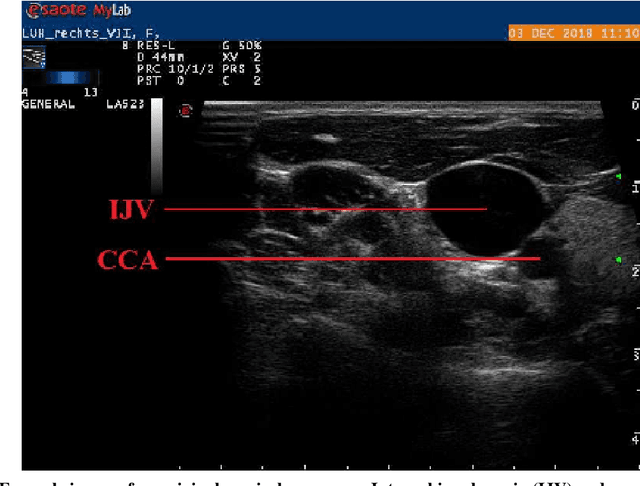
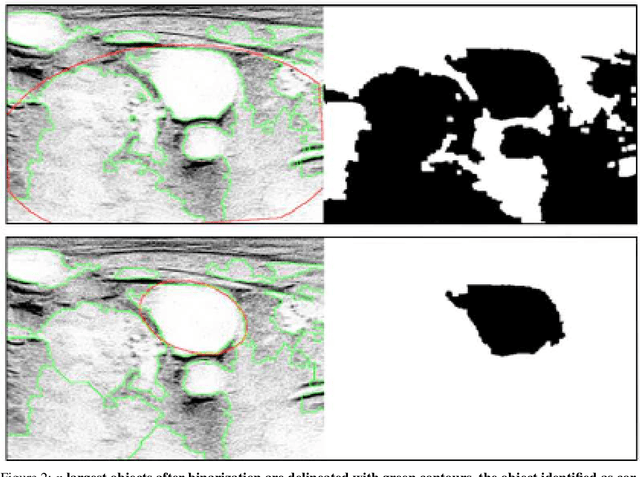
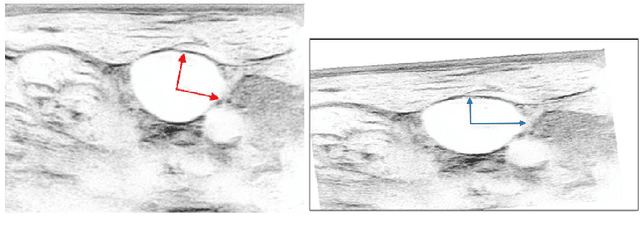
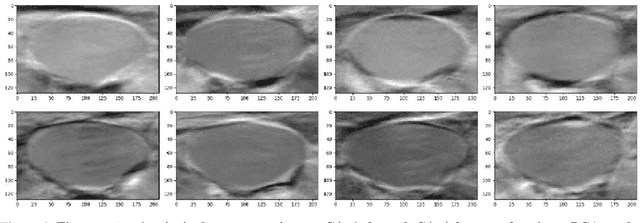
Abstract:Central venous catheters (CVC) are commonly inserted into the large veins of the neck, e.g. the internal jugular vein (IJV). CVC insertion may cause serious complications like misplacement into an artery or perforation of cervical vessels. Placing a CVC under sonographic guidance is an appropriate method to reduce such adverse events, if anatomical landmarks like venous and arterial vessels can be detected reliably. This task shall be solved by registration of patient individual images vs. an anatomically labelled reference image. In this work, a linear, affine transformation is performed on cervical sonograms, followed by a non-linear transformation to achieve a more precise registration. Voxelmorph (VM), a learning-based library for deformable image registration using a convolutional neural network (CNN) with U-Net structure was used for non-linear transformation. The impact of principal component analysis (PCA)-based pre-denoising of patient individual images, as well as the impact of modified net structures with differing complexities on registration results were examined visually and quantitatively, the latter using metrics for deformation and image similarity. Using the PCA-approximated cervical sonograms resulted in decreased mean deformation lengths between 18% and 66% compared to their original image counterparts, depending on net structure. In addition, reducing the number of convolutional layers led to improved image similarity with PCA images, while worsening in original images. Despite a large reduction of network parameters, no overall decrease in registration quality was observed, leading to the conclusion that the original net structure is oversized for the task at hand.
* 8 pages, 7 figures Presented at WSCG 2022
Estimation of mitral valve hinge point coordinates -- deep neural net for echocardiogram segmentation
Jan 20, 2023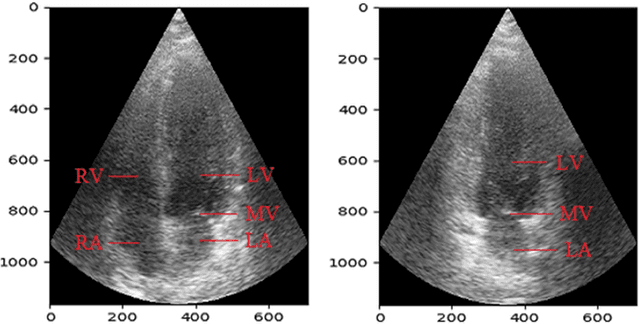
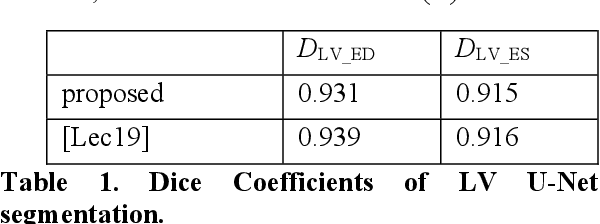
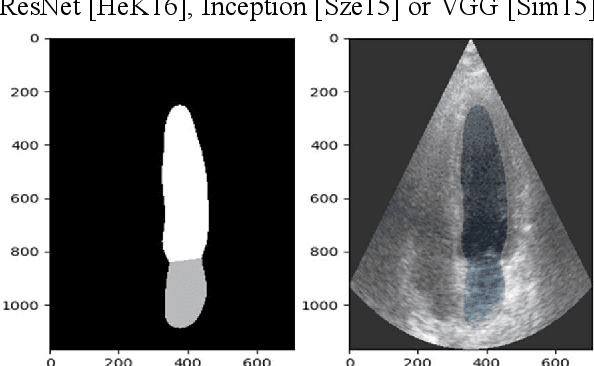
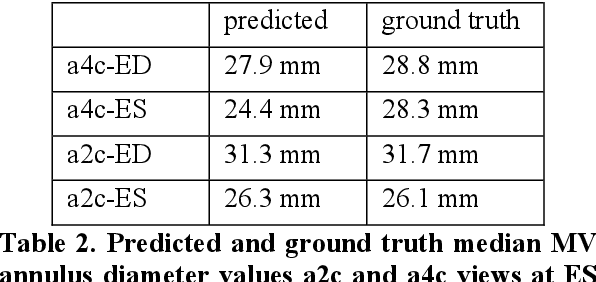
Abstract:Cardiac image segmentation is a powerful tool in regard to diagnostics and treatment of cardiovascular diseases. Purely feature-based detection of anatomical structures like the mitral valve is a laborious task due to specifically required feature engineering and is especially challenging in echocardiograms, because of their inherently low contrast and blurry boundaries between some anatomical structures. With the publication of further annotated medical datasets and the increase in GPU processing power, deep learning-based methods in medical image segmentation became more feasible in the past years. We propose a fully automatic detection method for mitral valve hinge points, which uses a U-Net based deep neural net to segment cardiac chambers in echocardiograms in a first step, and subsequently extracts the mitral valve hinge points from the resulting segmentations in a second step. Results measured with this automatic detection method were compared to reference coordinate values, which with median absolute hinge point coordinate errors of 1.35 mm for the x- (15-85 percentile range: [0.3 mm; 3.15 mm]) and 0.75 mm for the y- coordinate (15-85 percentile range: [0.15 mm; 1.88 mm]).
* 8 Pages, 11 figures Presented at WSCG 2022
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge