Harsh Verma
Comparing and combining some popular NER approaches on Biomedical tasks
May 30, 2023Abstract:We compare three simple and popular approaches for NER: 1) SEQ (sequence-labeling with a linear token classifier) 2) SeqCRF (sequence-labeling with Conditional Random Fields), and 3) SpanPred (span-prediction with boundary token embeddings). We compare the approaches on 4 biomedical NER tasks: GENIA, NCBI-Disease, LivingNER (Spanish), and SocialDisNER (Spanish). The SpanPred model demonstrates state-of-the-art performance on LivingNER and SocialDisNER, improving F1 by 1.3 and 0.6 F1 respectively. The SeqCRF model also demonstrates state-of-the-art performance on LivingNER and SocialDisNER, improving F1 by 0.2 F1 and 0.7 respectively. The SEQ model is competitive with the state-of-the-art on the LivingNER dataset. We explore some simple ways of combining the three approaches. We find that majority voting consistently gives high precision and high F1 across all 4 datasets. Lastly, we implement a system that learns to combine the predictions of SEQ and SpanPred, generating systems that consistently give high recall and high F1 across all 4 datasets. On the GENIA dataset, we find that our learned combiner system significantly boosts F1(+1.2) and recall(+2.1) over the systems being combined. We release all the well-documented code necessary to reproduce all systems at https://github.com/flyingmothman/bionlp.
CLaC at SemEval-2023 Task 2: Comparing Span-Prediction and Sequence-Labeling approaches for NER
May 05, 2023Abstract:This paper summarizes the CLaC submission for the MultiCoNER 2 task which concerns the recognition of complex, fine-grained named entities. We compare two popular approaches for NER, namely Sequence Labeling and Span Prediction. We find that our best Span Prediction system performs slightly better than our best Sequence Labeling system on test data. Moreover, we find that using the larger version of XLM RoBERTa significantly improves performance. Post-competition experiments show that Span Prediction and Sequence Labeling approaches improve when they use special input tokens (<s> and </s>) of XLM-RoBERTa. The code for training all models, preprocessing, and post-processing is available at https://github.com/harshshredding/semeval2023-multiconer-paper.
CLaCLab at SocialDisNER: Using Medical Gazetteers for Named-Entity Recognition of Disease Mentions in Spanish Tweets
Sep 13, 2022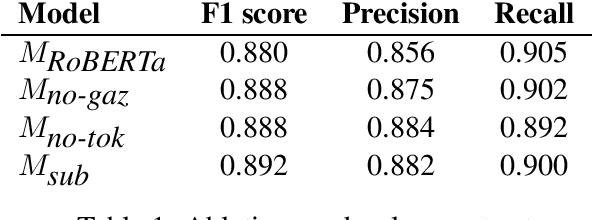
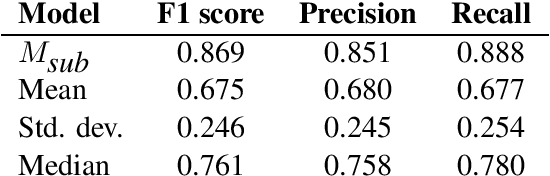
Abstract:This paper summarizes the CLaC submission for SMM4H 2022 Task 10 which concerns the recognition of diseases mentioned in Spanish tweets. Before classifying each token, we encode each token with a transformer encoder using features from Multilingual RoBERTa Large, UMLS gazetteer, and DISTEMIST gazetteer, among others. We obtain a strict F1 score of 0.869, with competition mean of 0.675, standard deviation of 0.245, and median of 0.761.
Convolutional Composer Classification
Nov 26, 2019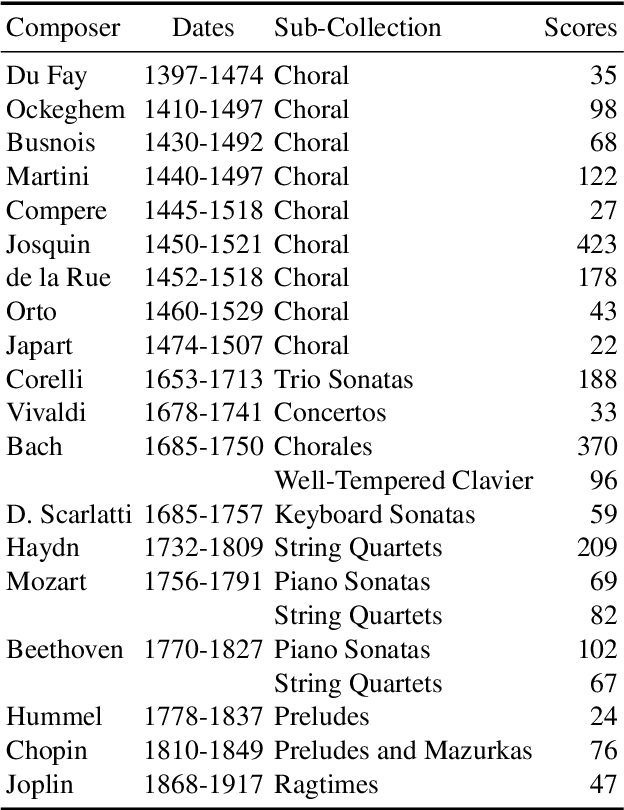
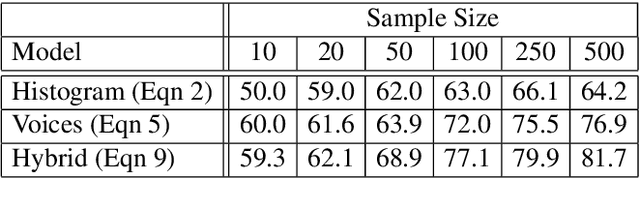
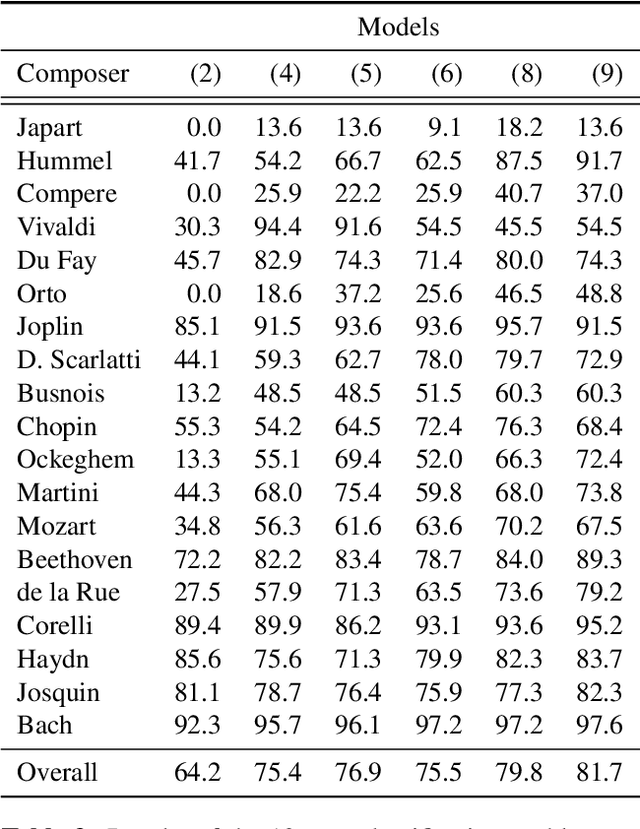
Abstract:This paper investigates end-to-end learnable models for attributing composers to musical scores. We introduce several pooled, convolutional architectures for this task and draw connections between our approach and classical learning approaches based on global and n-gram features. We evaluate models on a corpus of 2,500 scores from the KernScores collection, authored by a variety of composers spanning the Renaissance era to the early 20th century. This corpus has substantial overlap with the corpora used in several previous, smaller studies; we compare our results on subsets of the corpus to these previous works.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge