Guus Grimbergen
Evaluation of Multi-Slice Inputs to Convolutional Neural Networks for Medical Image Segmentation
Dec 22, 2019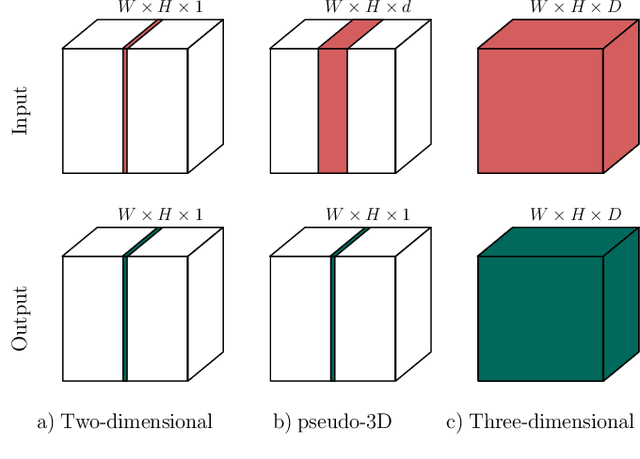
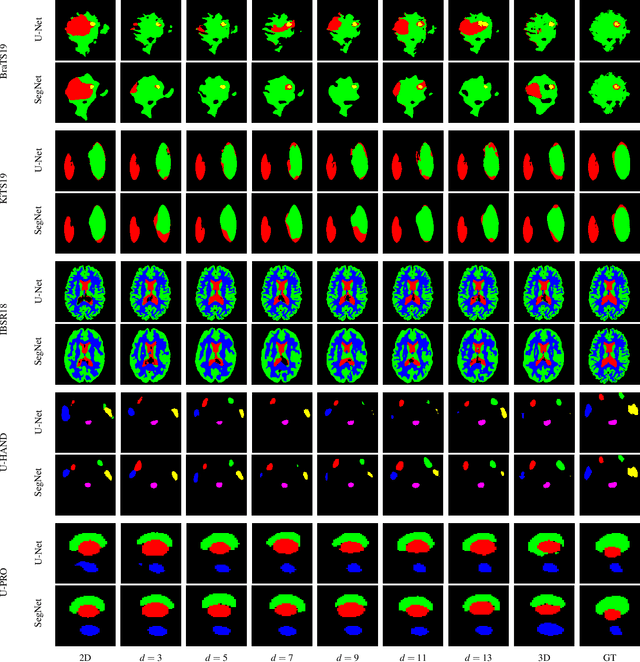
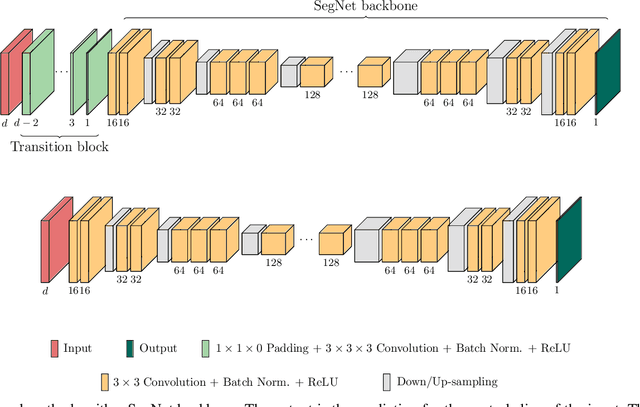
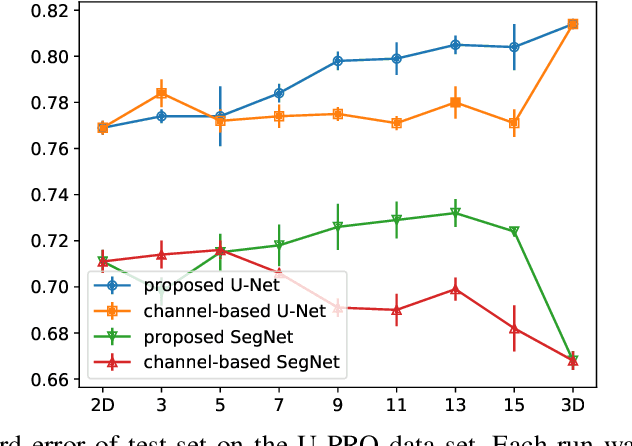
Abstract:When using Convolutional Neural Networks (CNNs) for segmentation of organs and lesions in medical images, the conventional approach is to work with inputs and outputs either as single slice (2D) or whole volumes (3D). One common alternative, in this study denoted as pseudo-3D, is to use a stack of adjacent slices as input and produce a prediction for at least the central slice. This approach gives the network the possibility to capture 3D spatial information, with only a minor additional computational cost. In this study, we systematically evaluate the segmentation performance and computational costs of this pseudo-3D approach as a function of the number of input slices, and compare the results to conventional end-to-end 2D and 3D CNNs. The standard pseudo-3D method regards the neighboring slices as multiple input image channels. We additionally evaluate a simple approach where the input stack is a volumetric input that is repeatably convolved in 3D to obtain a 2D feature map. This 2D map is in turn fed into a standard 2D network. We conducted experiments using two different CNN backbone architectures and on five diverse data sets covering different anatomical regions, imaging modalities, and segmentation tasks. We found that while both pseudo-3D methods can process a large number of slices at once and still be computationally much more efficient than fully 3D CNNs, a significant improvement over a regular 2D CNN was only observed for one of the five data sets. An analysis of the structural properties of the segmentation masks revealed no relations to the segmentation performance with respect to the number of input slices. The conclusion is therefore that in the general case, multi-slice inputs appear to not significantly improve segmentation results over using 2D or 3D CNNs.
End-to-End Cascaded U-Nets with a Localization Network for Kidney Tumor Segmentation
Oct 16, 2019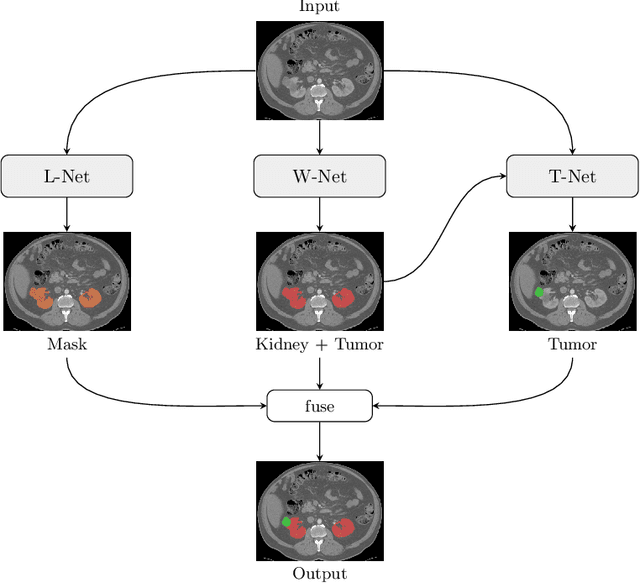
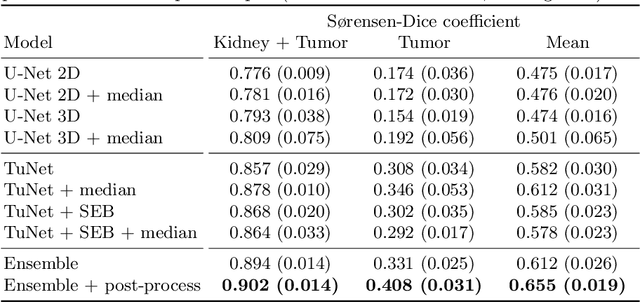
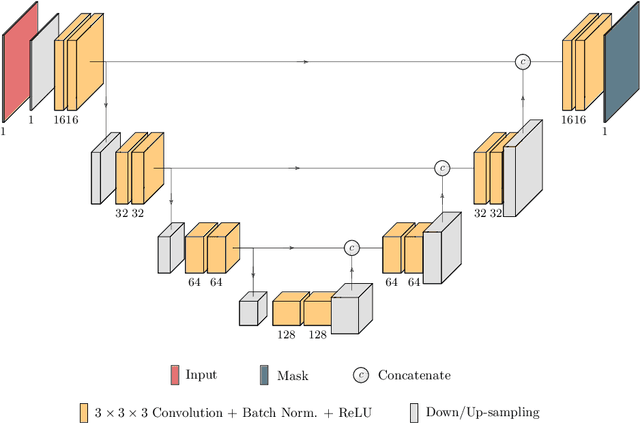
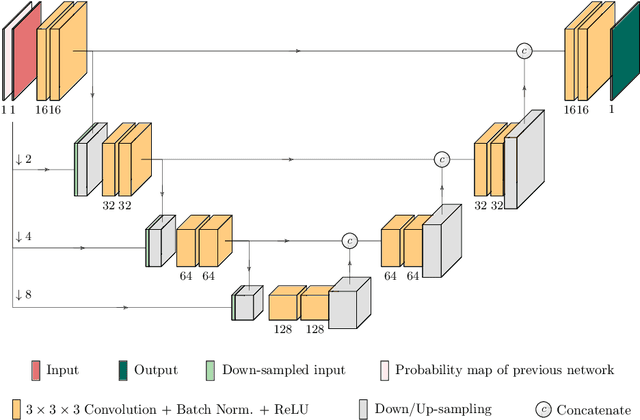
Abstract:Kidney tumor segmentation emerges as a new frontier of computer vision in medical imaging. This is partly due to its challenging manual annotation and great medical impact. Within the scope of the Kidney Tumor Segmentation Challenge 2019, that is aiming at combined kidney and tumor segmentation, this work proposes a novel combination of 3D U-Nets---collectively denoted TuNet---utilizing the resulting kidney masks for the consecutive tumor segmentation. The proposed method achieves a S{\o}rensen-Dice coefficient score of 0.902 for the kidney, and 0.408 for the tumor segmentation, computed from a five-fold cross-validation on the 210 patients available in the data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge