Gaetan Dissez
Predicting gene essentiality and drug response from perturbation screens in preclinical cancer models with LEAP: Layered Ensemble of Autoencoders and Predictors
Feb 21, 2025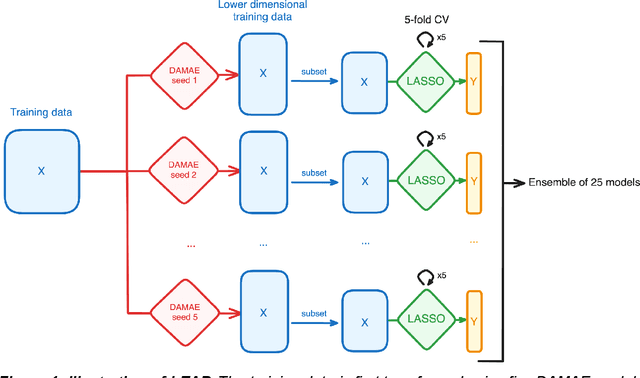
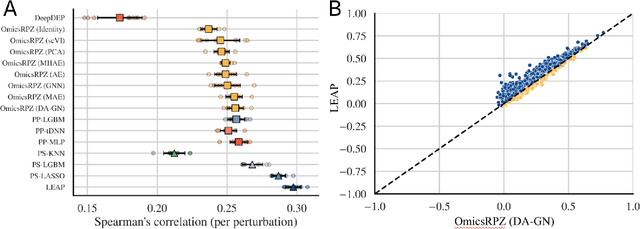
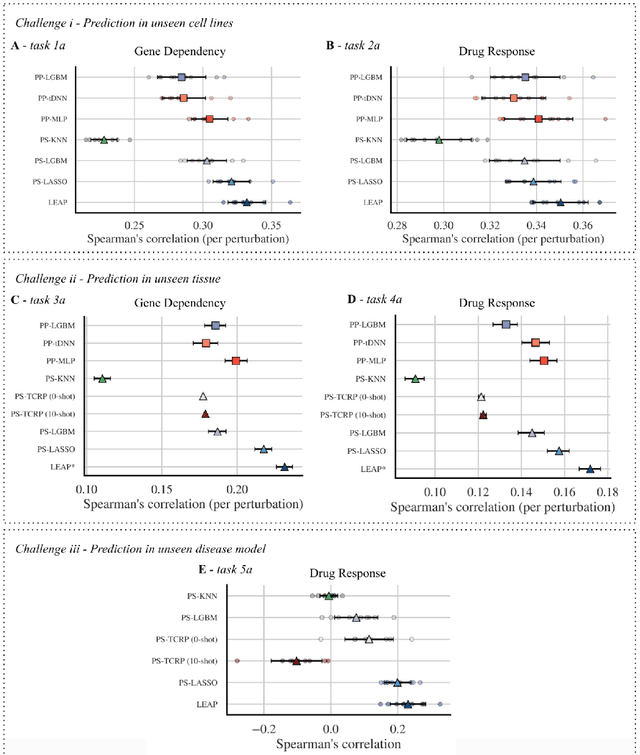
Abstract:Preclinical perturbation screens, where the effects of genetic, chemical, or environmental perturbations are systematically tested on disease models, hold significant promise for machine learning-enhanced drug discovery due to their scale and causal nature. Predictive models can infer perturbation responses for previously untested disease models based on molecular profiles. These in silico labels can expand databases and guide experimental prioritization. However, modelling perturbation-specific effects and generating robust prediction performances across diverse biological contexts remain elusive. We introduce LEAP (Layered Ensemble of Autoencoders and Predictors), a novel ensemble framework to improve robustness and generalization. LEAP leverages multiple DAMAE (Data Augmented Masked Autoencoder) representations and LASSO regressors. By combining diverse gene expression representation models learned from different random initializations, LEAP consistently outperforms state-of-the-art approaches in predicting gene essentiality or drug responses in unseen cell lines, tissues and disease models. Notably, our results show that ensembling representation models, rather than prediction models alone, yields superior predictive performance. Beyond its performance gains, LEAP is computationally efficient, requires minimal hyperparameter tuning and can therefore be readily incorporated into drug discovery pipelines to prioritize promising targets and support biomarker-driven stratification. The code and datasets used in this work are made publicly available.
Enhancing Early Lung Cancer Detection on Chest Radiographs with AI-assistance: A Multi-Reader Study
Aug 31, 2022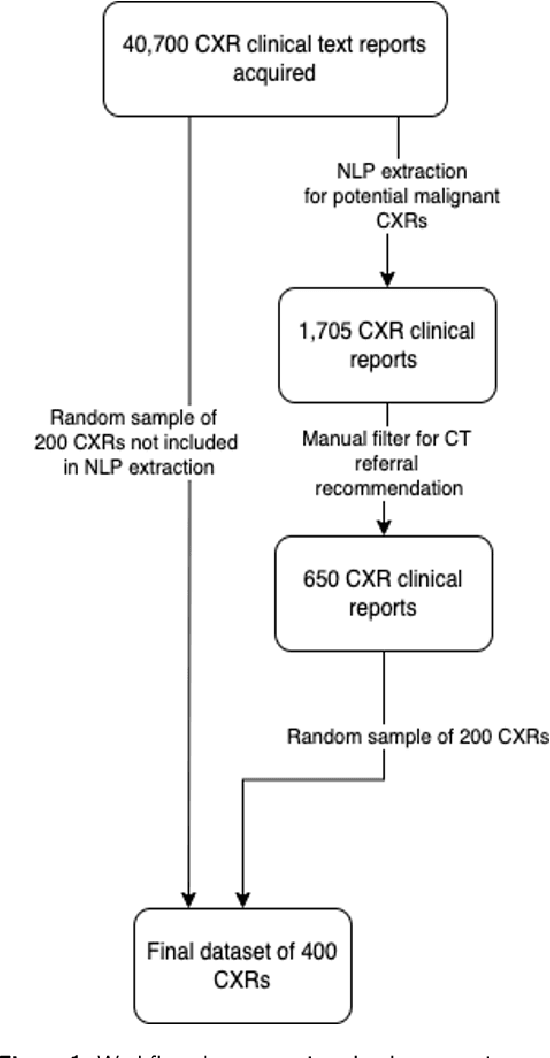
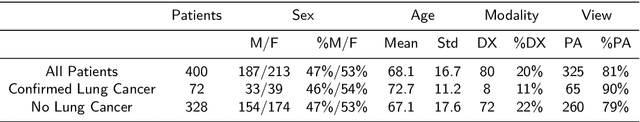
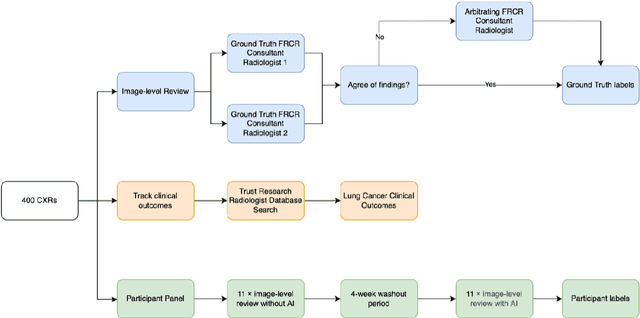
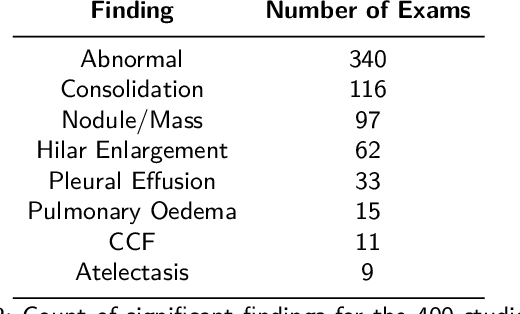
Abstract:Objectives: The present study evaluated the impact of a commercially available explainable AI algorithm in augmenting the ability of clinicians to identify lung cancer on chest X-rays (CXR). Design: This retrospective study evaluated the performance of 11 clinicians for detecting lung cancer from chest radiographs, with and without assistance from a commercially available AI algorithm (red dot, Behold.ai) that predicts suspected lung cancer from CXRs. Clinician performance was evaluated against clinically confirmed diagnoses. Setting: The study analysed anonymised patient data from an NHS hospital; the dataset consisted of 400 chest radiographs from adult patients (18 years and above) who had a CXR performed in 2020, with corresponding clinical text reports. Participants: A panel of readers consisting of 11 clinicians (consultant radiologists, radiologist trainees and reporting radiographers) participated in this study. Main outcome measures: Overall accuracy, sensitivity, specificity and precision for detecting lung cancer on CXRs by clinicians, with and without AI input. Agreement rates between clinicians and performance standard deviation were also evaluated, with and without AI input. Results: The use of the AI algorithm by clinicians led to an improved overall performance for lung tumour detection, achieving an overall increase of 17.4% of lung cancers being identified on CXRs which would have otherwise been missed, an overall increase in detection of smaller tumours, a 24% and 13% increased detection of stage 1 and stage 2 lung cancers respectively, and standardisation of clinician performance. Conclusions: This study showed great promise in the clinical utility of AI algorithms in improving early lung cancer diagnosis and promoting health equity through overall improvement in reader performances, without impacting downstream imaging resources.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge