Flavio Fenton
Rotor Localization and Phase Mapping of Cardiac Excitation Waves using Deep Neural Networks
Sep 22, 2021

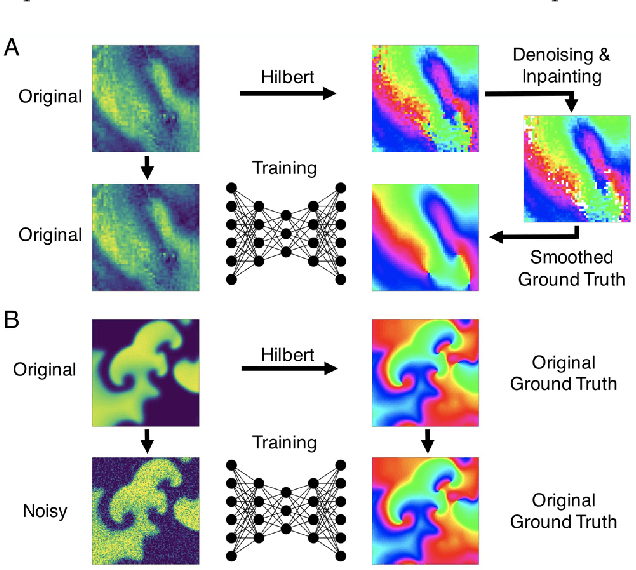
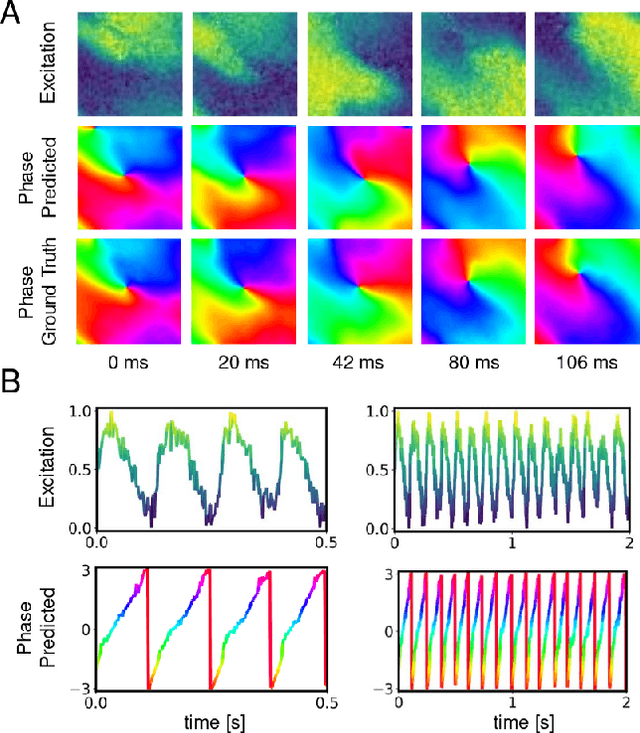
Abstract:The analysis of electrical impulse phenomena in cardiac muscle tissue is important for the diagnosis of heart rhythm disorders and other cardiac pathophysiology. Cardiac mapping techniques acquire numerous local temporal measurements and combine them to visualize the spread of electrophysiological wave phenomena across the heart surface. However, low spatial resolutions, sparse measurement locations, noise and other artifacts make it challenging to accurately visualize spatio-temporal activity. For instance, electro-anatomical catheter mapping is severely limited by the sparsity of the measurements and optical mapping is prone to noise and motion artifacts. In the past, several approaches have been proposed to obtain more reliable maps from noisy or sparse mapping data. Here, we demonstrate that deep learning can be used to compute phase maps and detect phase singularities from both noisy and sparse electrical mapping data with high precision and efficiency. The self-supervised deep learning approach is fundamentally different from classical phase mapping techniques. Rather than encoding a phase signal from time-series data, the network instead learns to directly associate short spatio-temporal sequences of electrical data with phase maps and the positions of phase singularities. Using this method, we were able to accurately compute phase maps and locate rotor cores even from extremely sparse and noisy data, generated from both optical mapping experiments and computer simulations. Neural networks are a promising alternative to conventional phase mapping and rotor core localization methods, that could be used in optical mapping studies in basic cardiovascular research as well as in the clinical setting for the analysis of atrial fibrillation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge