Federico Mattia Stefanini
Learning Bayesian Networks with Heterogeneous Agronomic Data Sets via Mixed-Effect Models and Hierarchical Clustering
Aug 29, 2023Abstract:Research involving diverse but related data sets, where associations between covariates and outcomes may vary, is prevalent in various fields including agronomic studies. In these scenarios, hierarchical models, also known as multilevel models, are frequently employed to assimilate information from different data sets while accommodating their distinct characteristics. However, their structure extend beyond simple heterogeneity, as variables often form complex networks of causal relationships. Bayesian networks (BNs) provide a powerful framework for modelling such relationships using directed acyclic graphs to illustrate the connections between variables. This study introduces a novel approach that integrates random effects into BN learning. Rooted in linear mixed-effects models, this approach is particularly well-suited for handling hierarchical data. Results from a real-world agronomic trial suggest that employing this approach enhances structural learning, leading to the discovery of new connections and the improvement of improved model specification. Furthermore, we observe a reduction in prediction errors from 28% to 17%. By extending the applicability of BNs to complex data set structures, this approach contributes to the effective utilisation of BNs for hierarchical agronomic data. This, in turn, enhances their value as decision-support tools in the field.
A probabilistic network for the diagnosis of acute cardiopulmonary diseases
May 12, 2018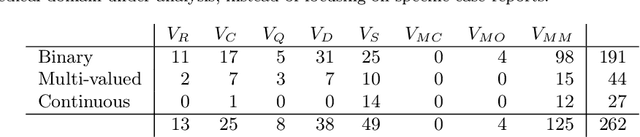

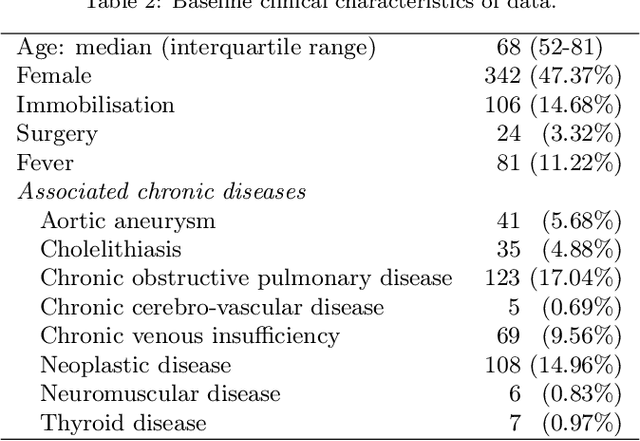

Abstract:In this paper, the development of a probabilistic network for the diagnosis of acute cardiopulmonary diseases is presented. This paper is a draft version of the article published after peer review in 2018 (https://doi.org/10.1002/bimj.201600206). A panel of expert physicians collaborated to specify the qualitative part, that is a directed acyclic graph defining a factorization of the joint probability distribution of domain variables. The quantitative part, that is the set of all conditional probability distributions defined by each factor, was estimated in the Bayesian paradigm: we applied a special formal representation, characterized by a low number of parameters and a parameterization intelligible for physicians, elicited the joint prior distribution of parameters from medical experts, and updated it by conditioning on a dataset of hospital patient records using Markov Chain Monte Carlo simulation. Refinement was cyclically performed until the probabilistic network provided satisfactory Concordance Index values for a selection of acute diseases and reasonable inference on six fictitious patient cases. The probabilistic network can be employed to perform medical diagnosis on a total of 63 diseases (38 acute and 25 chronic) on the basis of up to 167 patient findings.
* The DOI of the article published after peer review was added. A technical detail was added in Section 3.2, Formula 8 (as a consequence, the ID of all the subsequent formulas result augmented by 1 with respect to the previous version). The prior standard deviation of the Gamma distribution in Table 4 was fixed (in the previous version, the prior variance was indicated, instead)
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge