Evgeni Levin
Covered Information Disentanglement: Model Transparency via Unbiased Permutation Importance
Nov 21, 2021



Abstract:Model transparency is a prerequisite in many domains and an increasingly popular area in machine learning research. In the medical domain, for instance, unveiling the mechanisms behind a disease often has higher priority than the diagnostic itself since it might dictate or guide potential treatments and research directions. One of the most popular approaches to explain model global predictions is the permutation importance where the performance on permuted data is benchmarked against the baseline. However, this method and other related approaches will undervalue the importance of a feature in the presence of covariates since these cover part of its provided information. To address this issue, we propose Covered Information Disentanglement (CID), a method that considers all feature information overlap to correct the values provided by permutation importance. We further show how to compute CID efficiently when coupled with Markov random fields. We demonstrate its efficacy in adjusting permutation importance first on a controlled toy dataset and discuss its effect on real-world medical data.
Interpretable Models via Pairwise permutations algorithm
Nov 17, 2021

Abstract:One of the most common pitfalls often found in high dimensional biological data sets are correlations between the features. This may lead to statistical and machine learning methodologies overvaluing or undervaluing these correlated predictors, while the truly relevant ones are ignored. In this paper, we will define a new method called \textit{pairwise permutation algorithm} (PPA) with the aim of mitigating the correlation bias in feature importance values. Firstly, we provide a theoretical foundation, which builds upon previous work on permutation importance. PPA is then applied to a toy data set, where we demonstrate its ability to correct the correlation effect. We further test PPA on a microbiome shotgun dataset, to show that the PPA is already able to obtain biological relevant biomarkers.
Graph Space Embedding
Jul 31, 2019



Abstract:We propose the Graph Space Embedding (GSE), a technique that maps the input into a space where interactions are implicitly encoded, with little computations required. We provide theoretical results on an optimal regime for the GSE, namely a feasibility region for its parameters, and demonstrate the experimental relevance of our findings. Next, we introduce a strategy to gain insight on which interactions are responsible for the certain predictions, paving the way for a far more transparent model. In an empirical evaluation on a real-world clinical cohort containing patients with suspected coronary artery disease, the GSE achieves far better performance than traditional algorithms.
Classification of Quantitative Light-Induced Fluorescence Images Using Convolutional Neural Network
May 25, 2017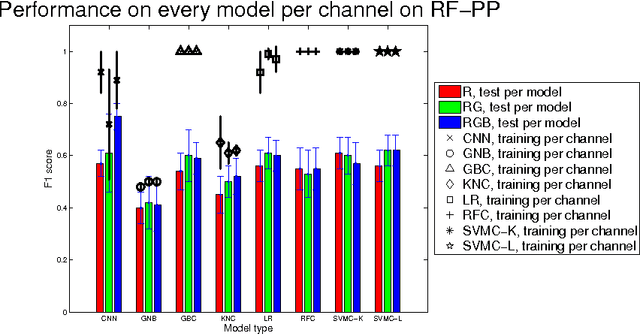
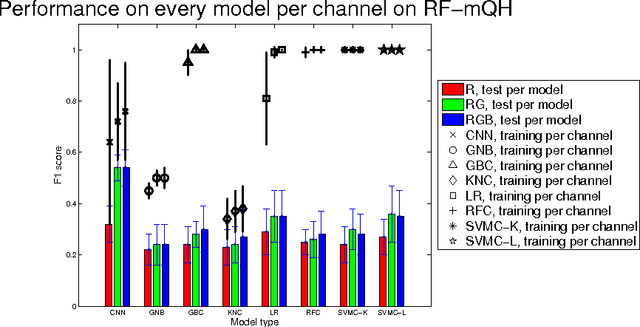
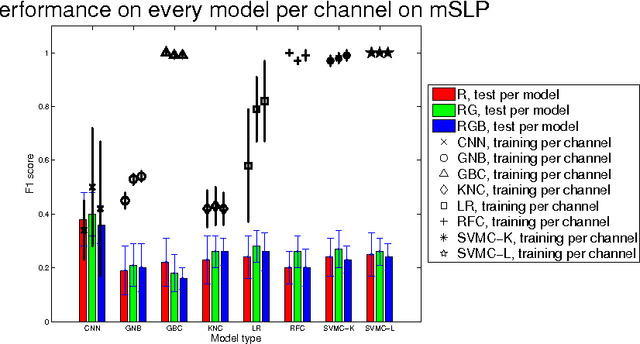
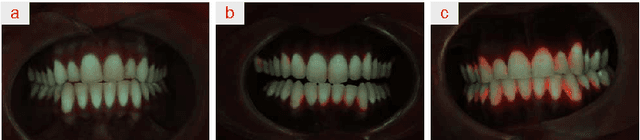
Abstract:Images are an important data source for diagnosis and treatment of oral diseases. The manual classification of images may lead to misdiagnosis or mistreatment due to subjective errors. In this paper an image classification model based on Convolutional Neural Network is applied to Quantitative Light-induced Fluorescence images. The deep neural network outperforms other state of the art shallow classification models in predicting labels derived from three different dental plaque assessment scores. The model directly benefits from multi-channel representation of the images resulting in improved performance when, besides the Red colour channel, additional Green and Blue colour channels are used.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge