Eugen Czeizler
Synergizing Data Imputation and Electronic Health Records for Advancing Prostate Cancer Research: Challenges, and Practical Applications
Oct 24, 2023Abstract:The presence of detailed clinical information in electronic health record (EHR) systems presents promising prospects for enhancing patient care through automated retrieval techniques. Nevertheless, it is widely acknowledged that accessing data within EHRs is hindered by various methodological challenges. Specifically, the clinical notes stored in EHRs are composed in a narrative form, making them prone to ambiguous formulations and highly unstructured data presentations, while structured reports commonly suffer from missing and/or erroneous data entries. This inherent complexity poses significant challenges when attempting automated large-scale medical knowledge extraction tasks, necessitating the application of advanced tools, such as natural language processing (NLP), as well as data audit techniques. This work aims to address these obstacles by creating and validating a novel pipeline designed to extract relevant data pertaining to prostate cancer patients. The objective is to exploit the inherent redundancies available within the integrated structured and unstructured data entries within EHRs in order to generate comprehensive and reliable medical databases, ready to be used in advanced research studies. Additionally, the study explores potential opportunities arising from these data, offering valuable prospects for advancing research in prostate cancer.
Identifying efficient controls of complex interaction networks using genetic algorithms
Jul 09, 2020
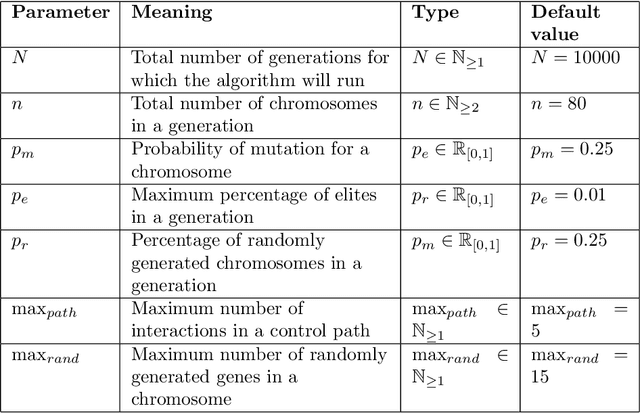
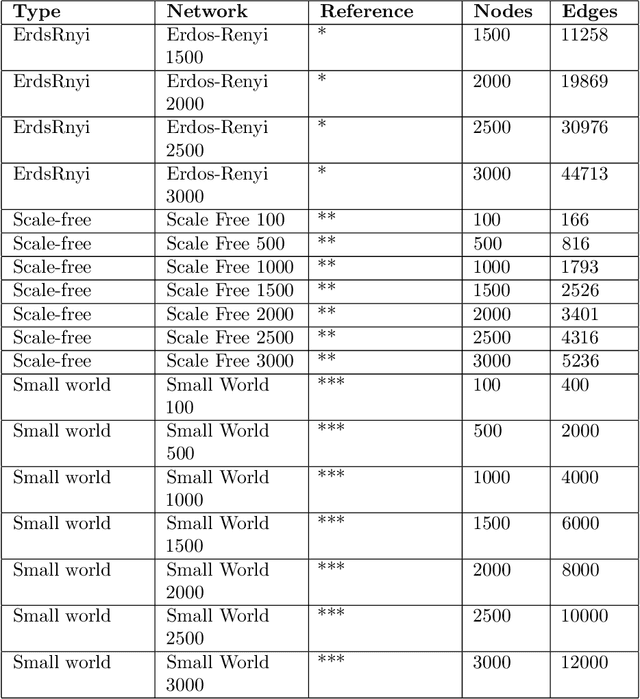
Abstract:Control theory has seen recently impactful applications in network science, especially in connections with applications in network medicine. A key topic of research is that of finding minimal external interventions that offer control over the dynamics of a given network, a problem known as network controllability. We propose in this article a new solution for this problem based on genetic algorithms. We tailor our solution for applications in computational drug repurposing, seeking to maximise its use of FDA-approved drug targets in a given disease-specific protein-protein interaction network. We show how our algorithm identifies a number of potentially efficient drugs for breast, ovarian, and pancreatic cancer. We demonstrate our algorithm on several benchmark networks from cancer medicine, social networks, electronic circuits, and several random networks with their edges distributed according to the Erd\H{o}s-R\'{e}nyi, the small-world, and the scale-free properties. Overall, we show that our new algorithm is more efficient in identifying relevant drug targets in a disease network, advancing the computational solutions needed for new therapeutic and drug repurposing approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge