Ethan Cerami
MatchMiner-AI: An Open-Source Solution for Cancer Clinical Trial Matching
Dec 23, 2024



Abstract:Clinical trials drive improvements in cancer treatments and outcomes. However, most adults with cancer do not participate in trials, and trials often fail to enroll enough patients to answer their scientific questions. Artificial intelligence could accelerate matching of patients to appropriate clinical trials. Here, we describe the development and evaluation of the MatchMiner-AI pipeline for clinical trial searching and ranking. MatchMiner-AI focuses on matching patients to potential trials based on core criteria describing clinical "spaces," or disease contexts, targeted by a trial. It aims to accelerate the human work of identifying potential matches, not to fully automate trial screening. The pipeline includes modules for extraction of key information from a patient's longitudinal electronic health record; rapid ranking of candidate trial-patient matches based on embeddings in vector space; and classification of whether a candidate match represents a reasonable clinical consideration. Code and synthetic data are available at https://huggingface.co/ksg-dfci/MatchMiner-AI . Model weights based on synthetic data are available at https://huggingface.co/ksg-dfci/TrialSpace and https://huggingface.co/ksg-dfci/TrialChecker . A simple cancer clinical trial search engine to demonstrate pipeline components is available at https://huggingface.co/spaces/ksg-dfci/trial_search_alpha .
MITI Minimum Information guidelines for highly multiplexed tissue images
Aug 21, 2021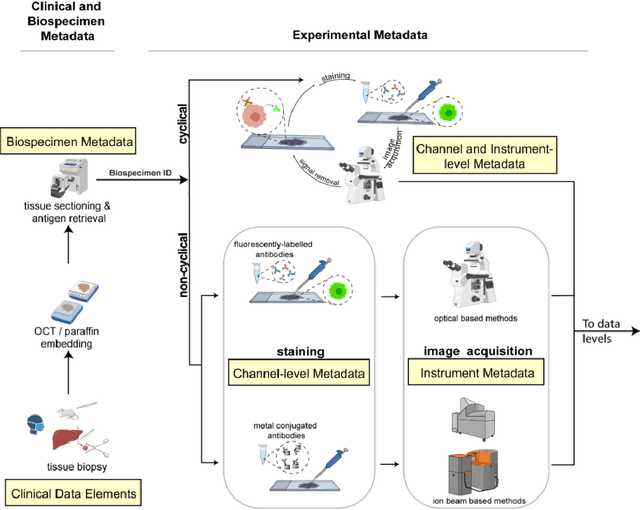
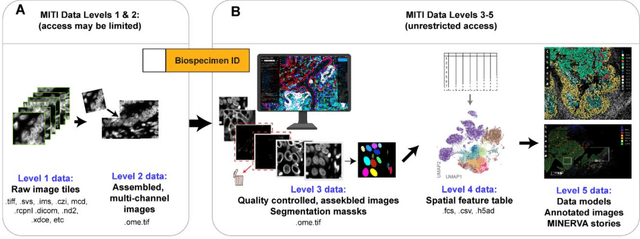
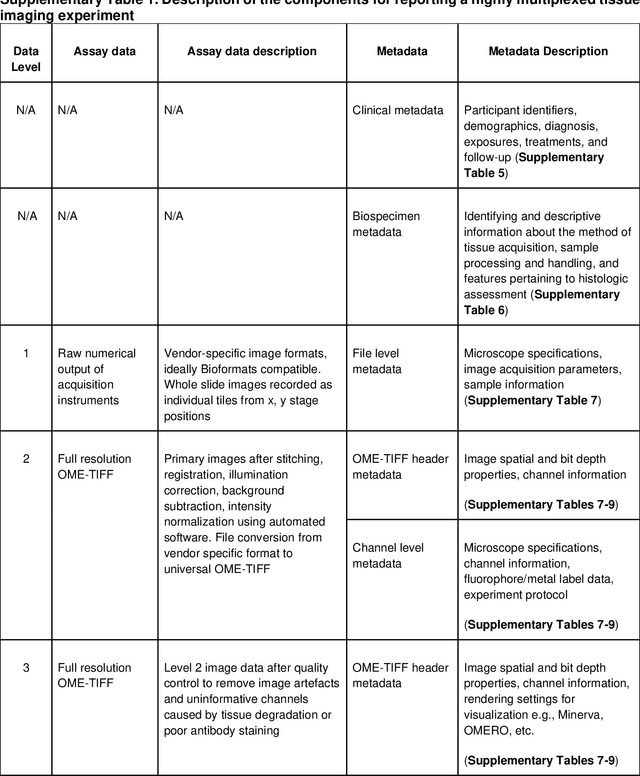
Abstract:The imminent release of atlases combining highly multiplexed tissue imaging with single cell sequencing and other omics data from human tissues and tumors creates an urgent need for data and metadata standards compliant with emerging and traditional approaches to histology. We describe the development of a Minimum Information about highly multiplexed Tissue Imaging (MITI) standard that draws on best practices from genomics and microscopy of cultured cells and model organisms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge