Emmanuel. M. Drakakis
Neuromorphic Processing: A Unifying Tutorial
Aug 13, 2021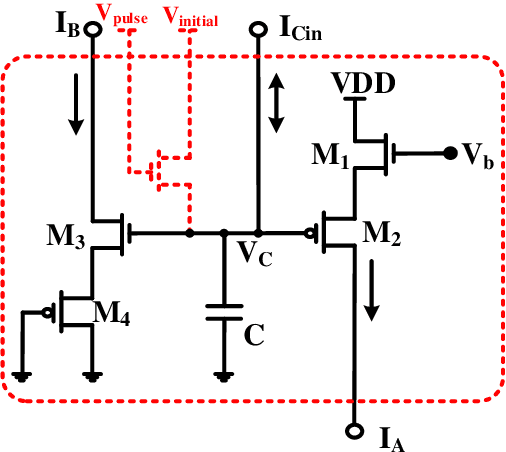
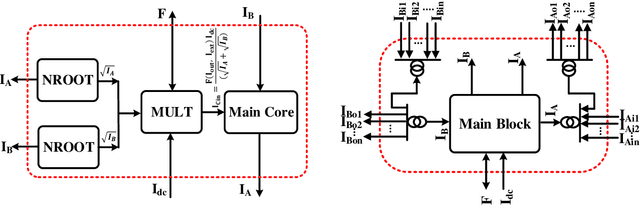
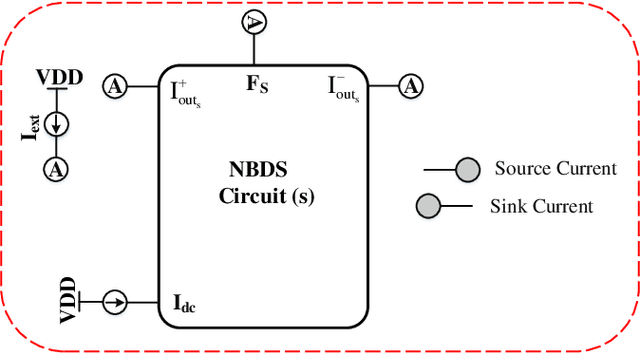
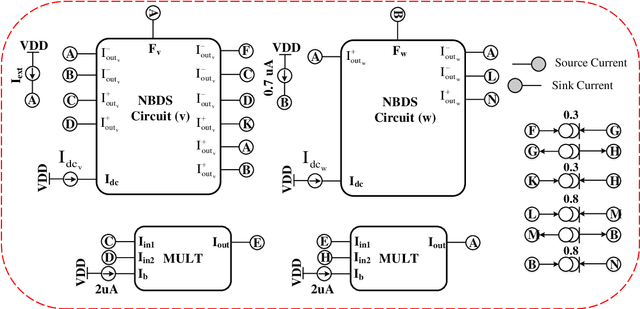
Abstract:All systolic or distributed neuromorphic architectures require power-efficient processing nodes. In this paper, a unifying tutorial is presented which implements multiple neuromorphic processing elements using a systematic analog approach including synapse, neuron and astrocyte models. It is shown that the proposed approach can successfully synthesize multidimensional dynamical systems into analog circuitry with minimum effort.
A Generalized Strong-Inversion CMOS Circuitry for Neuromorphic Applications
Jul 28, 2020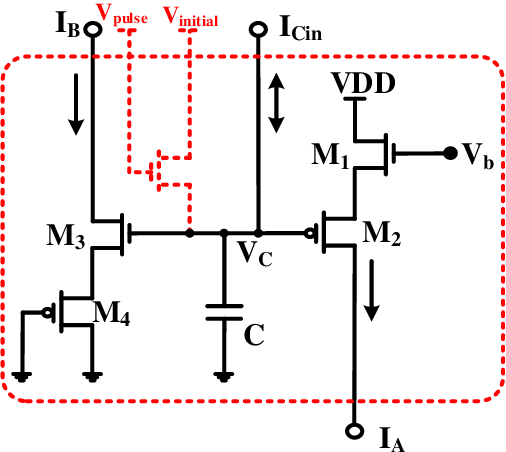
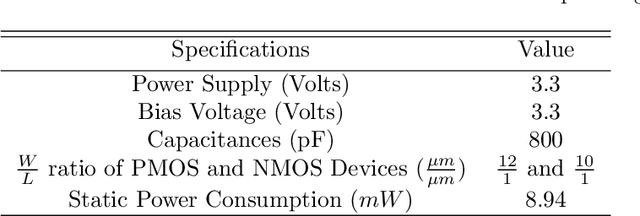
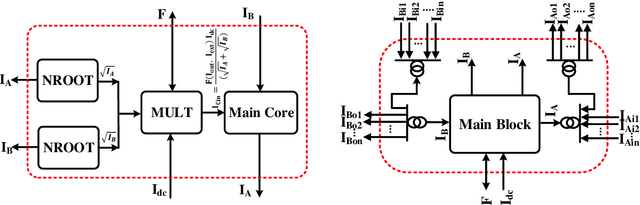
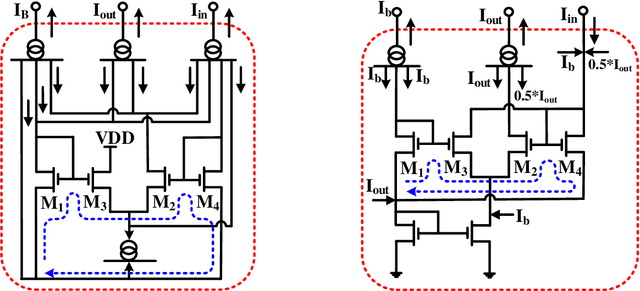
Abstract:It has always been a challenge in the neuromorphic field to systematically translate biological models into analog electronic circuitry. In this paper, a generalized circuit design platform is introduced where biological models can be conveniently implemented using CMOS circuitry operating in strong-inversion. The application of the method is demonstrated by synthesizing a relatively complex two-dimensional (2-D) nonlinear neuron model. The validity of our approach is verified by nominal simulated results with realistic process parameters from the commercially available AMS 0.35 um technology. The circuit simulation results exhibit regular spiking responses in good agreement with their mathematical counterpart.
A High GOPs/Slice Time Series Classifier for Portable and Embedded Biomedical Applications
Nov 01, 2018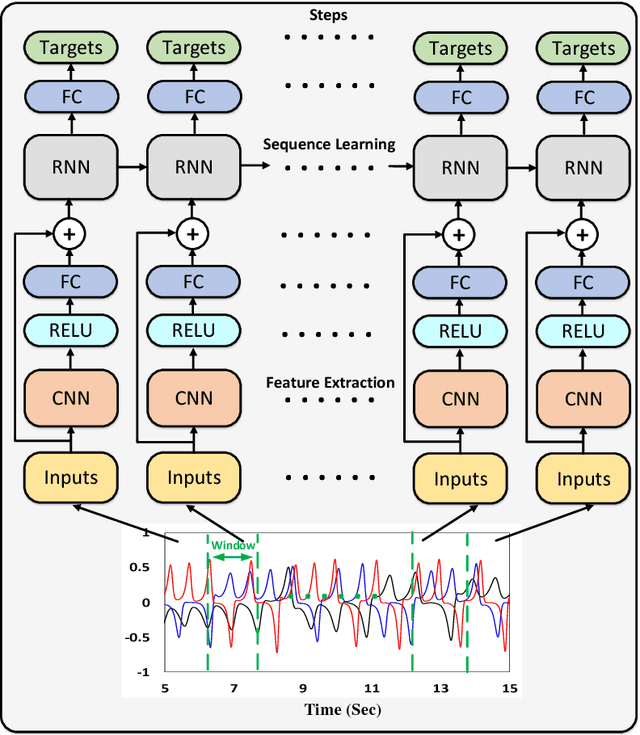

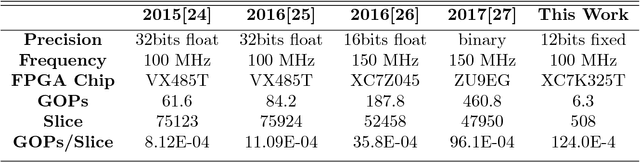
Abstract:Nowadays a diverse range of physiological data can be captured continuously for various applications in particular wellbeing and healthcare. Such data require efficient methods for classification and analysis. Deep learning algorithms have shown remarkable potential regarding such analyses, however, the use of these algorithms on low-power wearable devices is challenged by resource constraints such as area and power consumption. Most of the available on-chip deep learning processors contain complex and dense hardware architectures in order to achieve the highest possible throughput. Such a trend in hardware design may not be efficient in applications where on-node computation is required and the focus is more on the area and power efficiency as in the case of portable and embedded biomedical devices. This paper presents an efficient time-series classifier capable of automatically detecting effective features and classifying the input signals in real-time. In the proposed classifier, throughput is traded off with hardware complexity and cost using resource sharing techniques. A Convolutional Neural Network (CNN) is employed to extract input features and then a Long-Short-Term-Memory (LSTM) architecture with ternary weight precision classifies the input signals according to the extracted features. Hardware implementation on a Xilinx FPGA confirm that the proposed hardware can accurately classify multiple complex biomedical time series data with low area and power consumption and outperform all previously presented state-of-the-art records. Most notably, our classifier reaches 1.3$\times$ higher GOPs/Slice than similar state of the art FPGA-based accelerators.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge