Emmanuel K. Tsinda
Primer C-VAE: An interpretable deep learning primer design method to detect emerging virus variants
Mar 03, 2025Abstract:Motivation: PCR is more economical and quicker than Next Generation Sequencing for detecting target organisms, with primer design being a critical step. In epidemiology with rapidly mutating viruses, designing effective primers is challenging. Traditional methods require substantial manual intervention and struggle to ensure effective primer design across different strains. For organisms with large, similar genomes like Escherichia coli and Shigella flexneri, differentiating between species is also difficult but crucial. Results: We developed Primer C-VAE, a model based on a Variational Auto-Encoder framework with Convolutional Neural Networks to identify variants and generate specific primers. Using SARS-CoV-2, our model classified variants (alpha, beta, gamma, delta, omicron) with 98% accuracy and generated variant-specific primers. These primers appeared with >95% frequency in target variants and <5% in others, showing good performance in in-silico PCR tests. For Alpha, Delta, and Omicron, our primer pairs produced fragments <200 bp, suitable for qPCR detection. The model also generated effective primers for organisms with longer gene sequences like E. coli and S. flexneri. Conclusion: Primer C-VAE is an interpretable deep learning approach for developing specific primer pairs for target organisms. This flexible, semi-automated and reliable tool works regardless of sequence completeness and length, allowing for qPCR applications and can be applied to organisms with large and highly similar genomes.
Deep learning forward and reverse primer design to detect SARS-CoV-2 emerging variants
Sep 25, 2022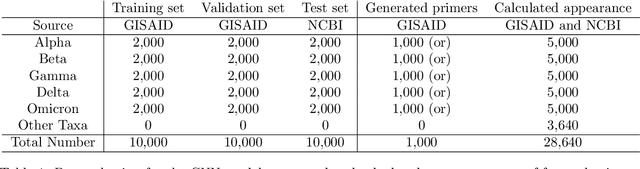
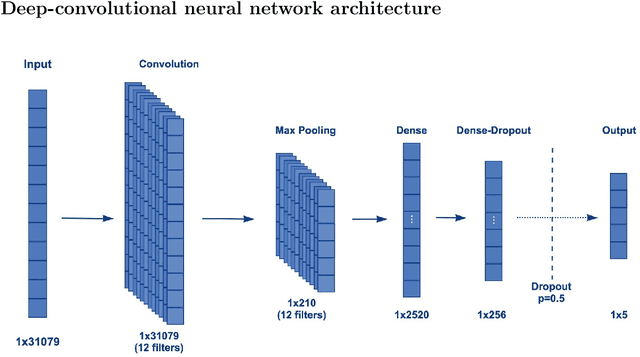
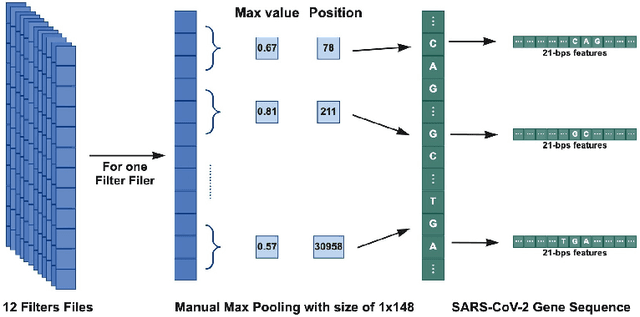
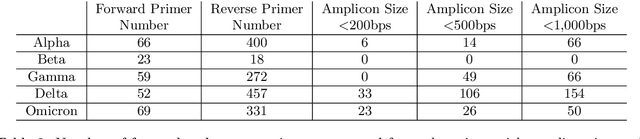
Abstract:Surges that have been observed at different periods in the number of COVID-19 cases are associated with the emergence of multiple SARS-CoV-2 (Severe Acute Respiratory Virus) variants. The design of methods to support laboratory detection are crucial in the monitoring of these variants. Hence, in this paper, we develop a semi-automated method to design both forward and reverse primer sets to detect SARS-CoV-2 variants. To proceed, we train deep Convolution Neural Networks (CNNs) to classify labelled SARS-CoV-2 variants and identify partial genomic features needed for the forward and reverse Polymerase Chain Reaction (PCR) primer design. Our proposed approach supplements existing ones while promoting the emerging concept of neural network assisted primer design for PCR. Our CNN model was trained using a database of SARS-CoV-2 full-length genomes from GISAID and tested on a separate dataset from NCBI, with 98\% accuracy for the classification of variants. This result is based on the development of three different methods of feature extraction, and the selected primer sequences for each SARS-CoV-2 variant detection (except Omicron) were present in more than 95 \% of sequences in an independent set of 5000 same variant sequences, and below 5 \% in other independent datasets with 5000 sequences of each variant. In total, we obtain 22 forward and reverse primer pairs with flexible length sizes (18-25 base pairs) with an expected amplicon length ranging between 42 and 3322 nucleotides. Besides the feature appearance, in-silico primer checks confirmed that the identified primer pairs are suitable for accurate SARS-CoV-2 variant detection by means of PCR tests.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge