Elizabeth Mouchet
Class-Guided Image-to-Image Diffusion: Cell Painting from Brightfield Images with Class Labels
Mar 29, 2023Abstract:Image-to-image reconstruction problems with free or inexpensive metadata in the form of class labels appear often in biological and medical image domains. Existing text-guided or style-transfer image-to-image approaches do not translate to datasets where additional information is provided as discrete classes. We introduce and implement a model which combines image-to-image and class-guided denoising diffusion probabilistic models. We train our model on a real-world dataset of microscopy images used for drug discovery, with and without incorporating metadata labels. By exploring the properties of image-to-image diffusion with relevant labels, we show that class-guided image-to-image diffusion can improve the meaningful content of the reconstructed images and outperform the unguided model in useful downstream tasks.
Self-Supervised Learning of Phenotypic Representations from Cell Images with Weak Labels
Sep 16, 2022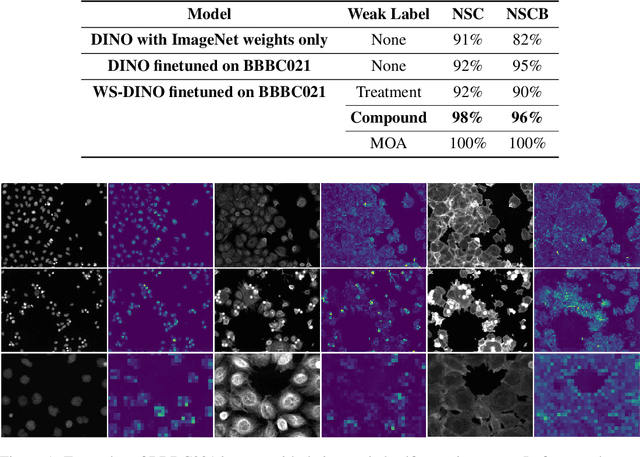
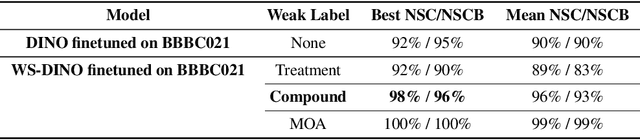
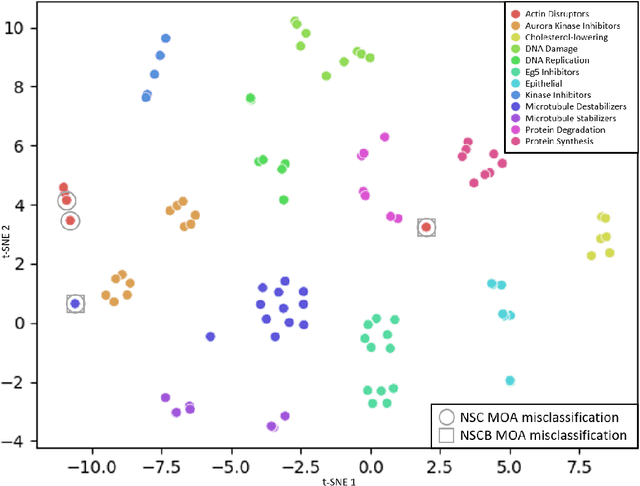
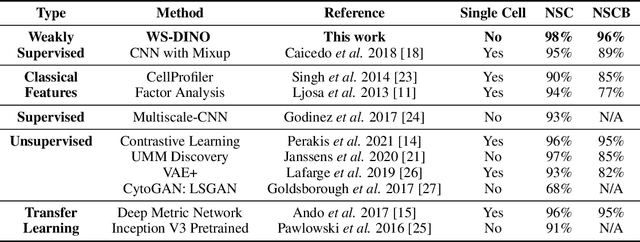
Abstract:We propose WS-DINO as a novel framework to use weak label information in learning phenotypic representations from high-content fluorescent images of cells. Our model is based on a knowledge distillation approach with a vision transformer backbone (DINO), and we use this as a benchmark model for our study. Using WS-DINO, we fine-tuned with weak label information available in high-content microscopy screens (treatment and compound), and achieve state-of-the-art performance in not-same-compound mechanism of action prediction on the BBBC021 dataset (98%), and not-same-compound-and-batch performance (96%) using the compound as the weak label. Our method bypasses single cell cropping as a pre-processing step, and using self-attention maps we show that the model learns structurally meaningful phenotypic profiles.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge