Elizabeth Cohen-Jonathan Moyal
CRCT, IUCT Oncopole
Multimodal Masked Autoencoder Pre-training for 3D MRI-Based Brain Tumor Analysis with Missing Modalities
May 01, 2025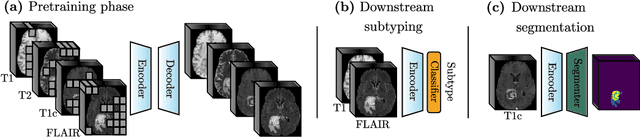
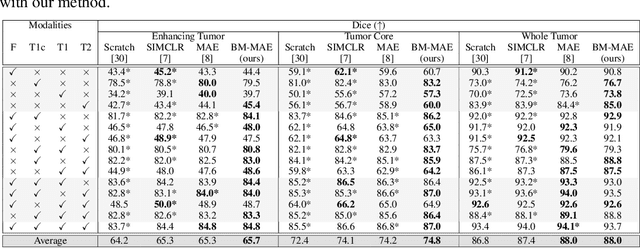
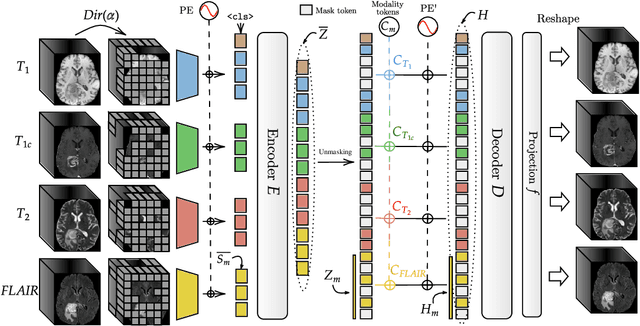
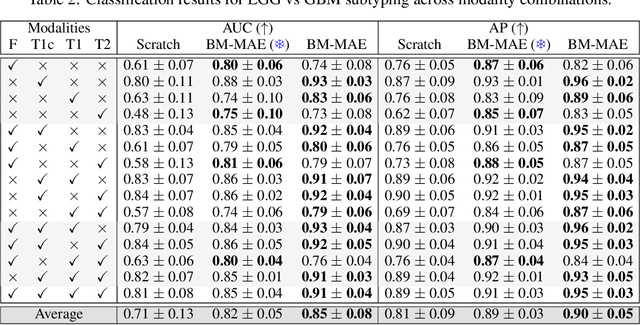
Abstract:Multimodal magnetic resonance imaging (MRI) constitutes the first line of investigation for clinicians in the care of brain tumors, providing crucial insights for surgery planning, treatment monitoring, and biomarker identification. Pre-training on large datasets have been shown to help models learn transferable representations and adapt with minimal labeled data. This behavior is especially valuable in medical imaging, where annotations are often scarce. However, applying this paradigm to multimodal medical data introduces a challenge: most existing approaches assume that all imaging modalities are available during both pre-training and fine-tuning. In practice, missing modalities often occur due to acquisition issues, specialist unavailability, or specific experimental designs on small in-house datasets. Consequently, a common approach involves training a separate model for each desired modality combination, making the process both resource-intensive and impractical for clinical use. Therefore, we introduce BM-MAE, a masked image modeling pre-training strategy tailored for multimodal MRI data. The same pre-trained model seamlessly adapts to any combination of available modalities, extracting rich representations that capture both intra- and inter-modal information. This allows fine-tuning on any subset of modalities without requiring architectural changes, while still benefiting from a model pre-trained on the full set of modalities. Extensive experiments show that the proposed pre-training strategy outperforms or remains competitive with baselines that require separate pre-training for each modality subset, while substantially surpassing training from scratch on several downstream tasks. Additionally, it can quickly and efficiently reconstruct missing modalities, highlighting its practical value. Code and trained models are available at: https://github.com/Lucas-rbnt/bmmae
Uncovering the Genetic Basis of Glioblastoma Heterogeneity through Multimodal Analysis of Whole Slide Images and RNA Sequencing Data
Oct 23, 2024Abstract:Glioblastoma is a highly aggressive form of brain cancer characterized by rapid progression and poor prognosis. Despite advances in treatment, the underlying genetic mechanisms driving this aggressiveness remain poorly understood. In this study, we employed multimodal deep learning approaches to investigate glioblastoma heterogeneity using joint image/RNA-seq analysis. Our results reveal novel genes associated with glioblastoma. By leveraging a combination of whole-slide images and RNA-seq, as well as introducing novel methods to encode RNA-seq data, we identified specific genetic profiles that may explain different patterns of glioblastoma progression. These findings provide new insights into the genetic mechanisms underlying glioblastoma heterogeneity and highlight potential targets for therapeutic intervention.
DRIM: Learning Disentangled Representations from Incomplete Multimodal Healthcare Data
Sep 25, 2024Abstract:Real-life medical data is often multimodal and incomplete, fueling the growing need for advanced deep learning models capable of integrating them efficiently. The use of diverse modalities, including histopathology slides, MRI, and genetic data, offers unprecedented opportunities to improve prognosis prediction and to unveil new treatment pathways. Contrastive learning, widely used for deriving representations from paired data in multimodal tasks, assumes that different views contain the same task-relevant information and leverages only shared information. This assumption becomes restrictive when handling medical data since each modality also harbors specific knowledge relevant to downstream tasks. We introduce DRIM, a new multimodal method for capturing these shared and unique representations, despite data sparsity. More specifically, given a set of modalities, we aim to encode a representation for each one that can be divided into two components: one encapsulating patient-related information common across modalities and the other, encapsulating modality-specific details. This is achieved by increasing the shared information among different patient modalities while minimizing the overlap between shared and unique components within each modality. Our method outperforms state-of-the-art algorithms on glioma patients survival prediction tasks, while being robust to missing modalities. To promote reproducibility, the code is made publicly available at https://github.com/Lucas-rbnt/DRIM
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge