Dorian Bagni
SynLlama: Generating Synthesizable Molecules and Their Analogs with Large Language Models
Mar 16, 2025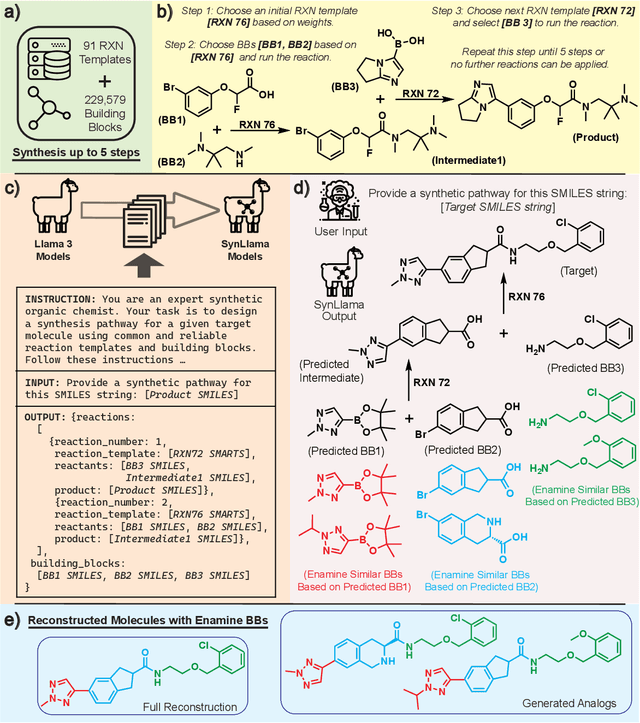
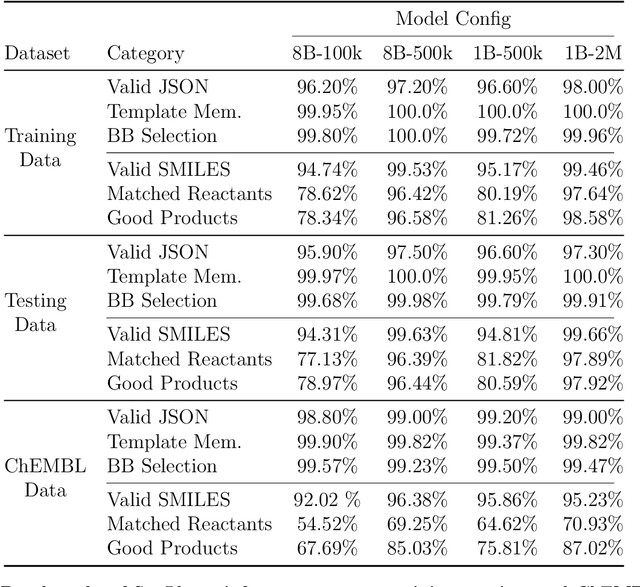
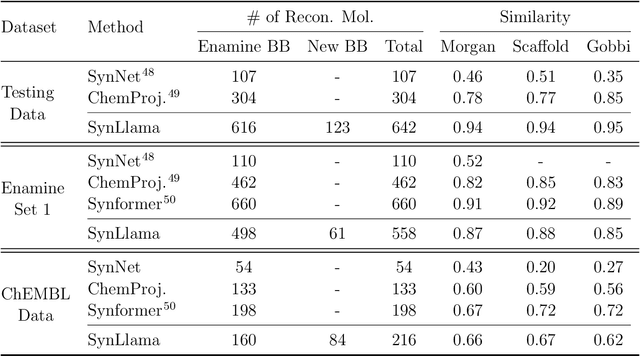
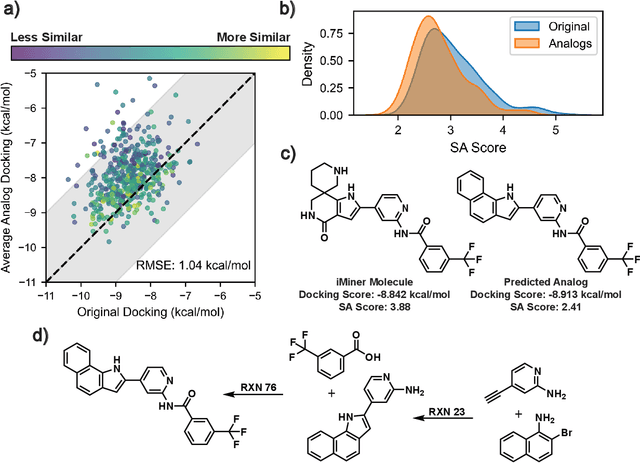
Abstract:Generative machine learning models for small molecule drug discovery have shown immense promise, but many molecules generated by this approach are too difficult to synthesize to be worth further investigation or further development. We present a novel approach by fine-tuning Meta's Llama3 large language models (LLMs) to create SynLlama, which generates full synthetic pathways made of commonly accessible Enamine building blocks and robust organic reaction templates. SynLlama explores a large synthesizable space using significantly less data compared to other state-of-the-art methods, and offers strong performance in bottom-up synthesis, synthesizable analog generation, and hit expansion, offering medicinal chemists a valuable tool for drug discovery developments. We find that SynLlama can effectively generalize to unseen yet purchasable building blocks, meaning that its reconstruction capabilities extend to a broader synthesizable chemical space than the training data.
SmileyLlama: Modifying Large Language Models for Directed Chemical Space Exploration
Sep 03, 2024



Abstract:Here we show that a Large Language Model (LLM) can serve as a foundation model for a Chemical Language Model (CLM) which performs at or above the level of CLMs trained solely on chemical SMILES string data. Using supervised fine-tuning (SFT) and direct preference optimization (DPO) on the open-source Llama LLM, we demonstrate that we can train an LLM to respond to prompts such as generating molecules with properties of interest to drug development. This overall framework allows an LLM to not just be a chatbot client for chemistry and materials tasks, but can be adapted to speak more directly as a CLM which can generate molecules with user-specified properties.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge