Dirk Drasdo
Diffusion-weighted MRI-guided needle biopsies permit quantitative tumor heterogeneity assessment and cell load estimation
Mar 01, 2021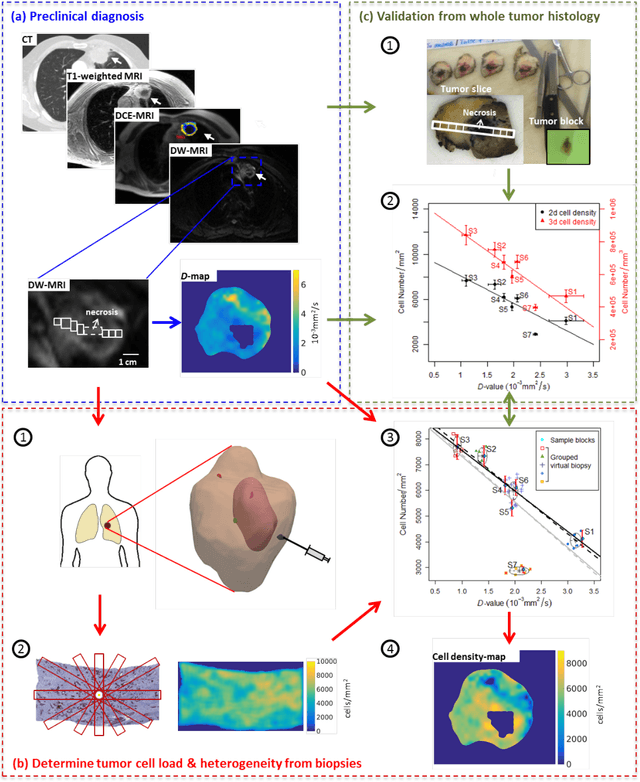
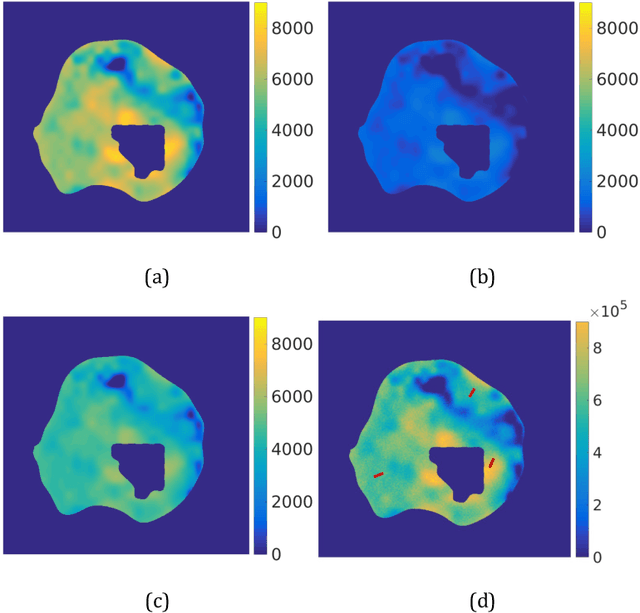
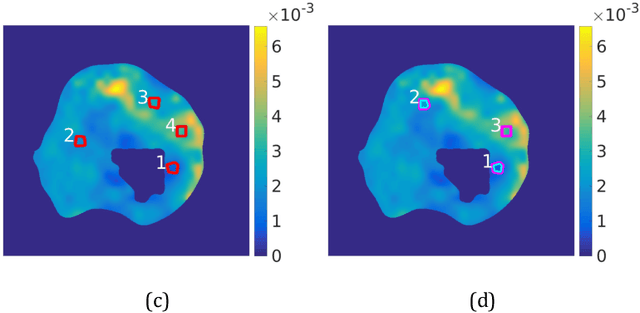
Abstract:Quantitative information on tumor heterogeneity and cell load could assist in designing effective and refined personalized treatment strategies. It was recently shown by us that such information can be inferred from the diffusion parameter D derived from the diffusion-weighted MRI (DWI) if a relation between D and cell density can be established. However, such relation cannot a priori be assumed to be constant for all patients and tumor types. Hence to assist in clinical decisions in palliative settings, the relation needs to be established without tumor resection. It is here demonstrated that biopsies may contain sufficient information for this purpose if the localization of biopsies is chosen as systematically elaborated in this paper. A superpixel-based method for automated optimal localization of biopsies from the DWI D-map is proposed. The performance of the DWI-guided procedure is evaluated by extensive simulations of biopsies. Needle biopsies yield sufficient histological information to establish a quantitative relationship between D-value and cell density, provided they are taken from regions with high, intermediate, and low D-value in DWI. The automated localization of the biopsy regions is demonstrated from a NSCLC patient tumor. In this case, even two or three biopsies give a reasonable estimate. Simulations of needle biopsies under different conditions indicate that the DWI-guidance highly improves the estimation results. Tumor cellularity and heterogeneity in solid tumors may be reliably investigated from DWI and a few needle biopsies that are sampled in regions of well-separated D-values, excluding adipose tissue. This procedure could provide a way of embedding in the clinical workflow assistance in cancer diagnosis and treatment based on personalized information.
Guided interactive image segmentation using machine learning and color based data set clustering
May 15, 2020Abstract:We present a novel approach that combines machine learning based interactive image segmentation with a two-stage clustering method for identification of similarly colored images enabling efficient batch image segmentation through guided reuse of interactively trained classifiers. The segmentation task is formulated as a supervised machine learning problem working on supervoxels. These visually homogeneous groups of voxels are characterized using local color, edge and texture features. Classifiers are interactively trained from sparse annotations in a iterative process of annotation refinement. Resulting models can be used for batch processing of previously unseen images. However, due to systemic discrepancies of image colorization classifier reusability is typically limited. By clustering a set of images into subsets of similar colorization, considering characteristic dominant color vectors obtained from the individual images it is possible to identify a minimal set of prototype images eligible for interactive segmentation. We demonstrate that limiting the reuse of pre-trained classifiers to images in the same color-cluster significantly improves the average segmentation performance of batch processing. The described methods are implemented in our free image processing and quantification software TiQuant released alongside this publication.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge