Dimitra Maoutsa
From geometry to dynamics: Learning overdamped Langevin dynamics from sparse observations with geometric constraints
Dec 29, 2025Abstract:How can we learn the laws underlying the dynamics of stochastic systems when their trajectories are sampled sparsely in time? Existing methods either require temporally resolved high-frequency observations, or rely on geometric arguments that apply only to conservative systems, limiting the range of dynamics they can recover. Here, we present a new framework that reconciles these two perspectives by reformulating inference as a stochastic control problem. Our method uses geometry-driven path augmentation, guided by the geometry in the system's invariant density to reconstruct likely trajectories and infer the underlying dynamics without assuming specific parametric models. Applied to overdamped Langevin systems, our approach accurately recovers stochastic dynamics even from extremely undersampled data, outperforming existing methods in synthetic benchmarks. This work demonstrates the effectiveness of incorporating geometric inductive biases into stochastic system identification methods.
Meta-learning three-factor plasticity rules for structured credit assignment with sparse feedback
Dec 11, 2025

Abstract:Biological neural networks learn complex behaviors from sparse, delayed feedback using local synaptic plasticity, yet the mechanisms enabling structured credit assignment remain elusive. In contrast, artificial recurrent networks solving similar tasks typically rely on biologically implausible global learning rules or hand-crafted local updates. The space of local plasticity rules capable of supporting learning from delayed reinforcement remains largely unexplored. Here, we present a meta-learning framework that discovers local learning rules for structured credit assignment in recurrent networks trained with sparse feedback. Our approach interleaves local neo-Hebbian-like updates during task execution with an outer loop that optimizes plasticity parameters via \textbf{tangent-propagation through learning}. The resulting three-factor learning rules enable long-timescale credit assignment using only local information and delayed rewards, offering new insights into biologically grounded mechanisms for learning in recurrent circuits.
Geometric constraints improve inference of sparsely observed stochastic dynamics
Apr 04, 2023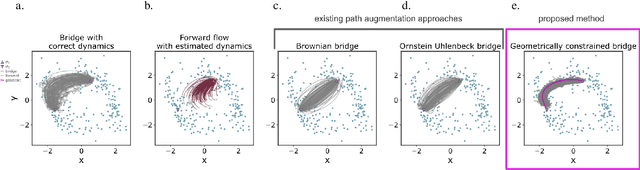
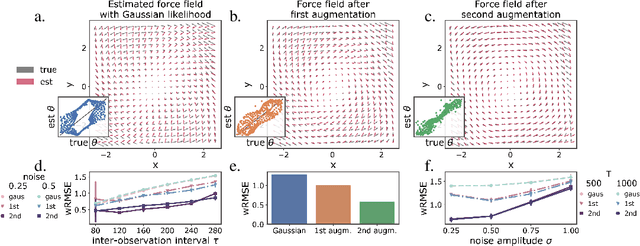
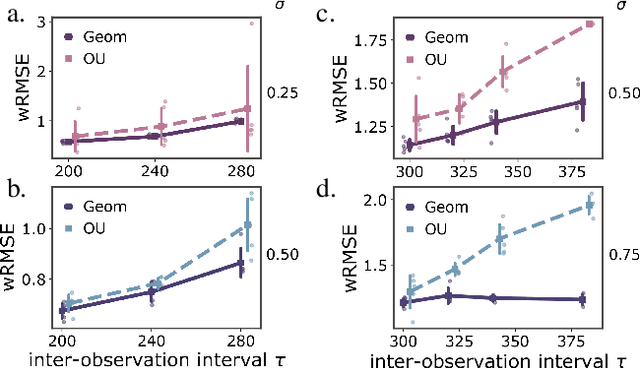
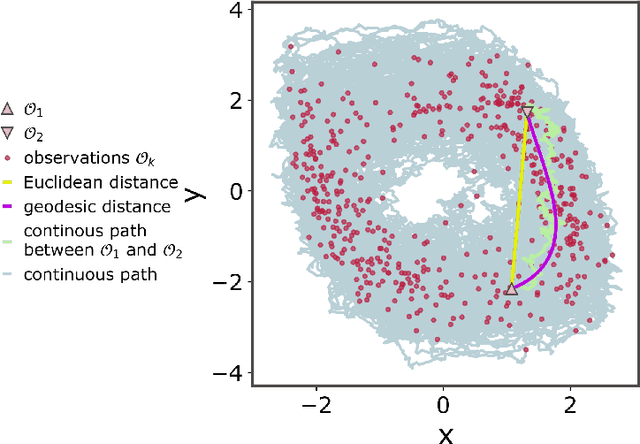
Abstract:The dynamics of systems of many degrees of freedom evolving on multiple scales are often modeled in terms of stochastic differential equations. Usually the structural form of these equations is unknown and the only manifestation of the system's dynamics are observations at discrete points in time. Despite their widespread use, accurately inferring these systems from sparse-in-time observations remains challenging. Conventional inference methods either focus on the temporal structure of observations, neglecting the geometry of the system's invariant density, or use geometric approximations of the invariant density, which are limited to conservative driving forces. To address these limitations, here, we introduce a novel approach that reconciles these two perspectives. We propose a path augmentation scheme that employs data-driven control to account for the geometry of the invariant system's density. Non-parametric inference on the augmented paths, enables efficient identification of the underlying deterministic forces of systems observed at low sampling rates.
Geometric path augmentation for inference of sparsely observed stochastic nonlinear systems
Jan 19, 2023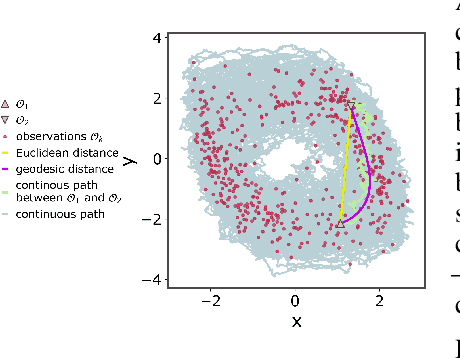
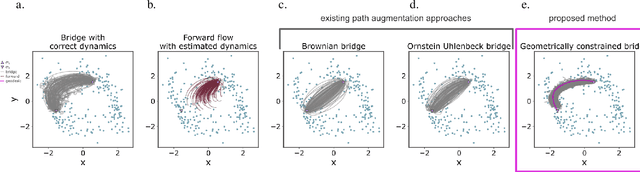
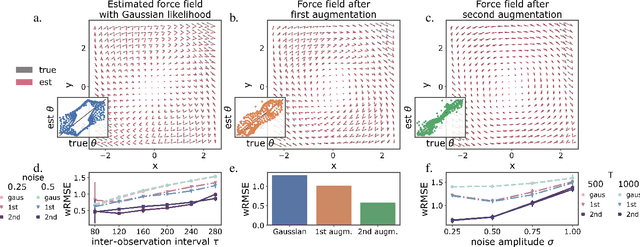
Abstract:Stochastic evolution equations describing the dynamics of systems under the influence of both deterministic and stochastic forces are prevalent in all fields of science. Yet, identifying these systems from sparse-in-time observations remains still a challenging endeavour. Existing approaches focus either on the temporal structure of the observations by relying on conditional expectations, discarding thereby information ingrained in the geometry of the system's invariant density; or employ geometric approximations of the invariant density, which are nevertheless restricted to systems with conservative forces. Here we propose a method that reconciles these two paradigms. We introduce a new data-driven path augmentation scheme that takes the local observation geometry into account. By employing non-parametric inference on the augmented paths, we can efficiently identify the deterministic driving forces of the underlying system for systems observed at low sampling rates.
Inferring network connectivity from event timing patterns
Mar 29, 2018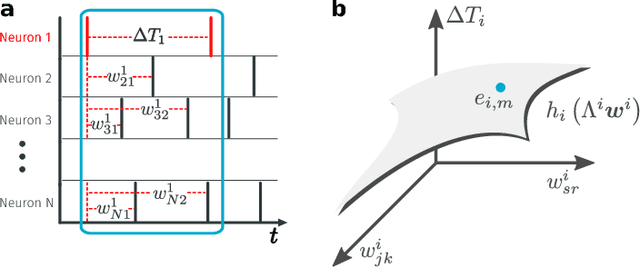
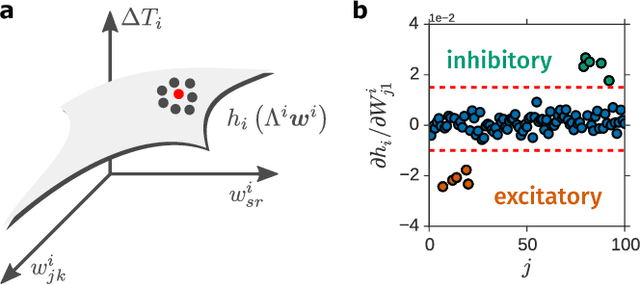
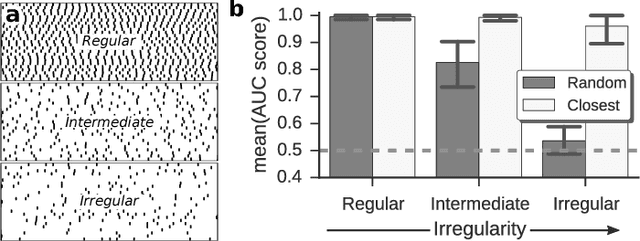
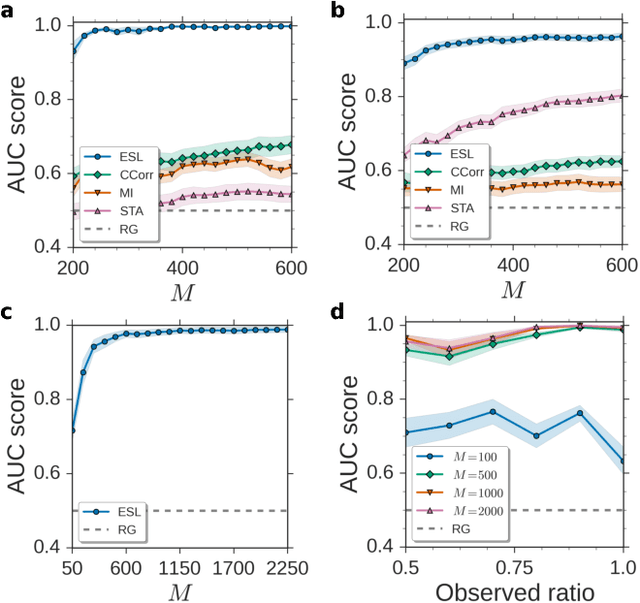
Abstract:Reconstructing network connectivity from the collective dynamics of a system typically requires access to its complete continuous-time evolution although these are often experimentally inaccessible. Here we propose a theory for revealing physical connectivity of networked systems only from the event time series their intrinsic collective dynamics generate. Representing the patterns of event timings in an event space spanned by inter-event and cross-event intervals, we reveal which other units directly influence the inter-event times of any given unit. For illustration, we linearize an event space mapping constructed from the spiking patterns in model neural circuits to reveal the presence or absence of synapses between any pair of neurons as well as whether the coupling acts in an inhibiting or activating (excitatory) manner. The proposed model-independent reconstruction theory is scalable to larger networks and may thus play an important role in the reconstruction of networks from biology to social science and engineering.
* 6 pages, 5 figures, The first two authors contributed equally to this paper, and should be regarded as co-first authors. [v2: metadata update]
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge