David S. Smith
MSTT-199: MRI Dataset for Musculoskeletal Soft Tissue Tumor Segmentation
Sep 04, 2024



Abstract:Accurate musculoskeletal soft tissue tumor segmentation is vital for assessing tumor size, location, diagnosis, and response to treatment, thereby influencing patient outcomes. However, segmentation of these tumors requires clinical expertise, and an automated segmentation model would save valuable time for both clinician and patient. Training an automatic model requires a large dataset of annotated images. In this work, we describe the collection of an MR imaging dataset of 199 musculoskeletal soft tissue tumors from 199 patients. We trained segmentation models on this dataset and then benchmarked them on a publicly available dataset. Our model achieved the state-of-the-art dice score of 0.79 out of the box without any fine tuning, which shows the diversity and utility of our curated dataset. We analyzed the model predictions and found that its performance suffered on fibrous and vascular tumors due to their diverse anatomical location, size, and intensity heterogeneity. The code and models are available in the following github repository, https://github.com/Reasat/mstt
Data Efficient Contrastive Learning in Histopathology using Active Sampling
Apr 03, 2023
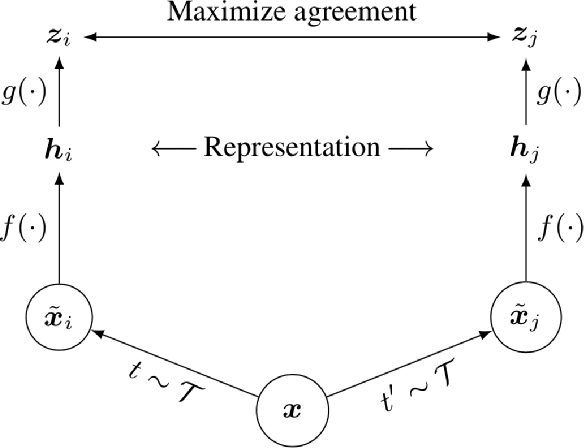
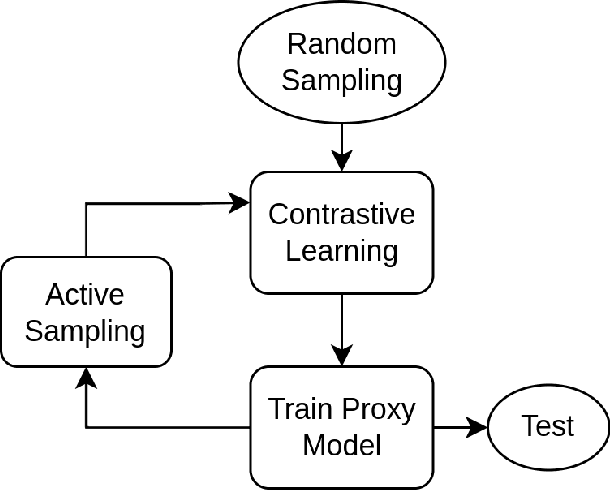
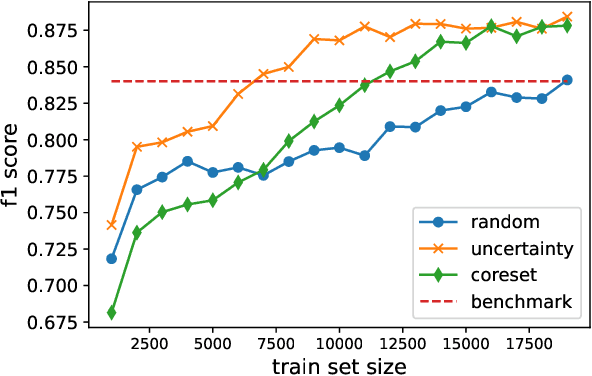
Abstract:Deep Learning based diagnostics systems can provide accurate and robust quantitative analysis in digital pathology. These algorithms require large amounts of annotated training data which is impractical in pathology due to the high resolution of histopathological images. Hence, self-supervised methods have been proposed to learn features using ad-hoc pretext tasks. The self-supervised training process is time consuming and often leads to subpar feature representation due to a lack of constrain on the learnt feature space, particularly prominent under data imbalance. In this work, we propose to actively sample the training set using a handful of labels and a small proxy network, decreasing sample requirement by 93% and training time by 99%.
RawArray: A Simple, Fast, and Extensible Archival Format for Numeric Data
Nov 30, 2021



Abstract:Raw data sizes are growing and proliferating in scientific research, driven by the success of data-hungry computational methods, such as machine learning. The preponderance of proprietary and shoehorned data formats make computations slower and make it harder to reproduce research and to port methods to new platforms. Here we present the RawArray format: a simple, fast, and extensible format for archival storage of multidimensional numeric arrays on disk. The RawArray file format is a simple concatenation of a header array and a data array. The header comprises seven or more 64-bit unsigned integers. The array data can be anything. Arbitrary user metadata can be appended to an RawArray file if desired, for example to store measurement details, color palettes, or geolocation data. We present benchmarks showing a factor of 2--3$\times$ speedup over HDF5 for a range of array sizes and a speedup of up to 20$\times$ in reading the common deep learning datasets MNIST and CIFAR10.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge