Daniela Zoeller
Combining propensity score methods with variational autoencoders for generating synthetic data in presence of latent sub-groups
Dec 12, 2023Abstract:In settings requiring synthetic data generation based on a clinical cohort, e.g., due to data protection regulations, heterogeneity across individuals might be a nuisance that we need to control or faithfully preserve. The sources of such heterogeneity might be known, e.g., as indicated by sub-groups labels, or might be unknown and thus reflected only in properties of distributions, such as bimodality or skewness. We investigate how such heterogeneity can be preserved and controlled when obtaining synthetic data from variational autoencoders (VAEs), i.e., a generative deep learning technique that utilizes a low-dimensional latent representation. To faithfully reproduce unknown heterogeneity reflected in marginal distributions, we propose to combine VAEs with pre-transformations. For dealing with known heterogeneity due to sub-groups, we complement VAEs with models for group membership, specifically from propensity score regression. The evaluation is performed with a realistic simulation design that features sub-groups and challenging marginal distributions. The proposed approach faithfully recovers the latter, compared to synthetic data approaches that focus purely on marginal distributions. Propensity scores add complementary information, e.g., when visualized in the latent space, and enable sampling of synthetic data with or without sub-group specific characteristics. We also illustrate the proposed approach with real data from an international stroke trial that exhibits considerable distribution differences between study sites, in addition to bimodality. These results indicate that describing heterogeneity by statistical approaches, such as propensity score regression, might be more generally useful for complementing generative deep learning for obtaining synthetic data that faithfully reflects structure from clinical cohorts.
Adapting deep generative approaches for getting synthetic data with realistic marginal distributions
May 14, 2021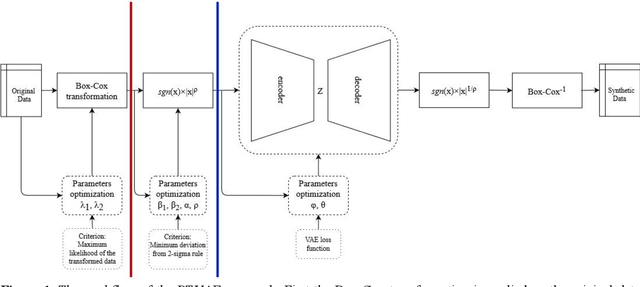
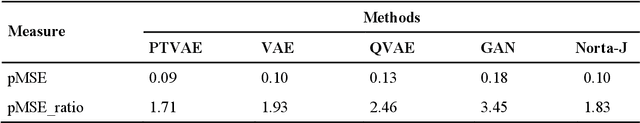
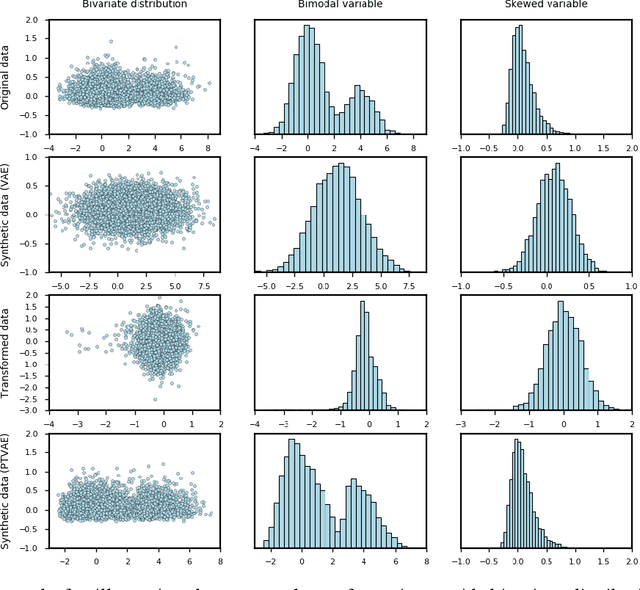
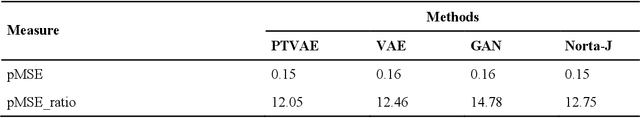
Abstract:Synthetic data generation is of great interest in diverse applications, such as for privacy protection. Deep generative models, such as variational autoencoders (VAEs), are a popular approach for creating such synthetic datasets from original data. Despite the success of VAEs, there are limitations when it comes to the bimodal and skewed marginal distributions. These deviate from the unimodal symmetric distributions that are encouraged by the normality assumption typically used for the latent representations in VAEs. While there are extensions that assume other distributions for the latent space, this does not generally increase flexibility for data with many different distributions. Therefore, we propose a novel method, pre-transformation variational autoencoders (PTVAEs), to specifically address bimodal and skewed data, by employing pre-transformations at the level of original variables. Two types of transformations are used to bring the data close to a normal distribution by a separate parameter optimization for each variable in a dataset. We compare the performance of our method with other state-of-the-art methods for synthetic data generation. In addition to the visual comparison, we use a utility measurement for a quantitative evaluation. The results show that the PTVAE approach can outperform others in both bimodal and skewed data generation. Furthermore, the simplicity of the approach makes it usable in combination with other extensions of VAE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge