Clément Mantoux
ARAMIS
Geometry-Aware Hamiltonian Variational Auto-Encoder
Oct 22, 2020
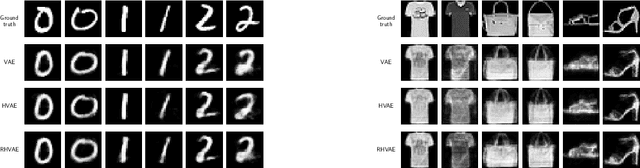
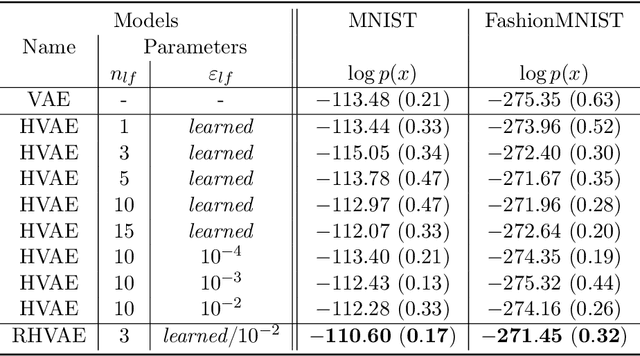
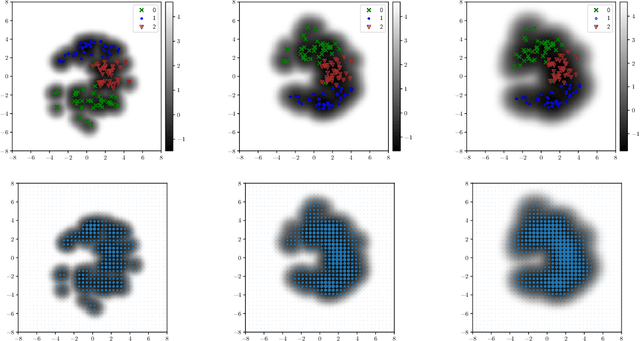
Abstract:Variational auto-encoders (VAEs) have proven to be a well suited tool for performing dimensionality reduction by extracting latent variables lying in a potentially much smaller dimensional space than the data. Their ability to capture meaningful information from the data can be easily apprehended when considering their capability to generate new realistic samples or perform potentially meaningful interpolations in a much smaller space. However, such generative models may perform poorly when trained on small data sets which are abundant in many real-life fields such as medicine. This may, among others, come from the lack of structure of the latent space, the geometry of which is often under-considered. We thus propose in this paper to see the latent space as a Riemannian manifold endowed with a parametrized metric learned at the same time as the encoder and decoder networks. This metric is then used in what we called the Riemannian Hamiltonian VAE which extends the Hamiltonian VAE introduced by arXiv:1805.11328 to better exploit the underlying geometry of the latent space. We argue that such latent space modelling provides useful information about its underlying structure leading to far more meaningful interpolations, more realistic data-generation and more reliable clustering.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge