Chun-An Chou
H2G2-Net: A Hierarchical Heterogeneous Graph Generative Network Framework for Discovery of Multi-Modal Physiological Responses
Jan 05, 2024Abstract:Discovering human cognitive and emotional states using multi-modal physiological signals draws attention across various research applications. Physiological responses of the human body are influenced by human cognition and commonly used to analyze cognitive states. From a network science perspective, the interactions of these heterogeneous physiological modalities in a graph structure may provide insightful information to support prediction of cognitive states. However, there is no clue to derive exact connectivity between heterogeneous modalities and there exists a hierarchical structure of sub-modalities. Existing graph neural networks are designed to learn on non-hierarchical homogeneous graphs with pre-defined graph structures; they failed to learn from hierarchical, multi-modal physiological data without a pre-defined graph structure. To this end, we propose a hierarchical heterogeneous graph generative network (H2G2-Net) that automatically learns a graph structure without domain knowledge, as well as a powerful representation on the hierarchical heterogeneous graph in an end-to-end fashion. We validate the proposed method on the CogPilot dataset that consists of multi-modal physiological signals. Extensive experiments demonstrate that our proposed method outperforms the state-of-the-art GNNs by 5%-20% in prediction accuracy.
Early ICU Mortality Prediction and Survival Analysis for Respiratory Failure
Sep 06, 2021



Abstract:Respiratory failure is the one of major causes of death in critical care unit. During the outbreak of COVID-19, critical care units experienced an extreme shortage of mechanical ventilation because of respiratory failure related syndromes. To help this, the early mortality risk prediction in patients who suffer respiratory failure can provide timely support for clinical treatment and resource management. In the study, we propose a dynamic modeling approach for early mortality risk prediction of the respiratory failure patients based on the first 24 hours ICU physiological data. Our proposed model is validated on the eICU collaborate database. We achieved a high AUROC performance (80-83%) and significantly improved AUCPR 4% on Day 5 since ICU admission, compared to the state-of-art prediction models. In addition, we illustrated that the survival curve includes the time-varying information for the early ICU admission survival analysis.
Mixed-Integer Optimization Approach to Learning Association Rules for Unplanned ICU Transfer
Aug 02, 2019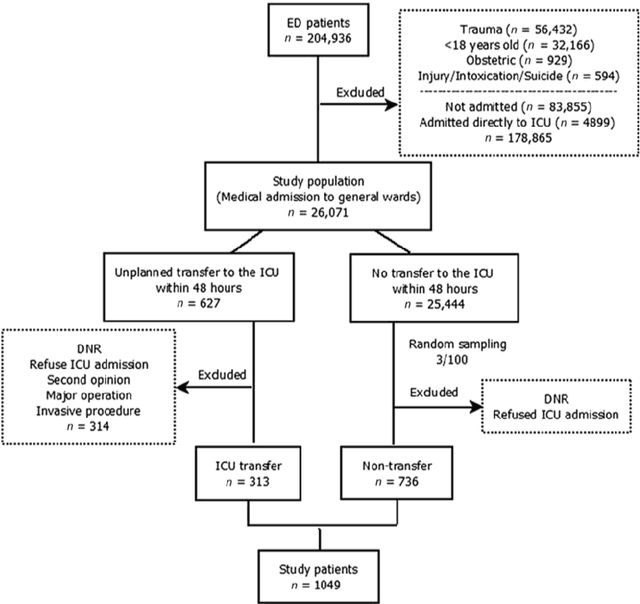


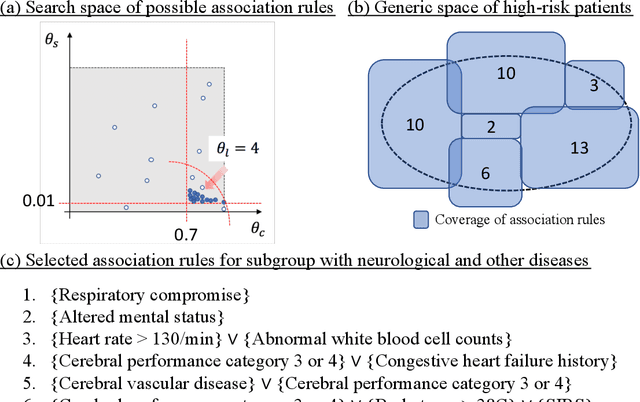
Abstract:After admission to emergency department (ED), patients with critical illnesses are transferred to intensive care unit (ICU) due to unexpected clinical deterioration occurrence. Identifying such unplanned ICU transfers is urgently needed for medical physicians to achieve two-fold goals: improving critical care quality and preventing mortality. A priority task is to understand the crucial rationale behind diagnosis results of individual patients during stay in ED, which helps prepare for an early transfer to ICU. Most existing prediction studies were based on univariate analysis or multiple logistic regression to provide one-size-fit-all results. However, patient condition varying from case to case may not be accurately examined by the only judgment. In this study, we present a new decision tool using a mathematical optimization approach aiming to automatically discover rules associating diagnostic features with high-risk outcome (i.e., unplanned transfers) in different deterioration scenarios. We consider four mutually exclusive patient subgroups based on the principal reasons of ED visits: infections, cardiovascular/respiratory diseases, gastrointestinal diseases, and neurological/other diseases at a suburban teaching hospital. The analysis results demonstrate significant rules associated with unplanned transfer outcome for each subgroups and also show comparable prediction accuracy, compared to state-of-the-art machine learning methods while providing easy-to-interpret symptom-outcome information.
Self-Expressive Subspace Clustering to Recognize Motion Dynamics of a Multi-Joint Coordination for Chronic Ankle Instability
Jan 06, 2019



Abstract:Ankle sprains and instability are major public health concerns. Up to 70% of individuals do not fully recover from a single ankle sprain and eventually develop chronic ankle instability (CAI). The diagnosis of CAI has been mainly based on self-report rather than objective biomechanical measures. The goal of this study is to quantitatively recognize the motion pattern of a multi-joint coordination using biosensor data from bilateral hip, knee, and ankle joints, and further distinguish between CAI and healthy cohorts. We propose an analytic framework, where a nonlinear subspace clustering method is developed to learn the motion dynamic patterns from an inter-connected network of multiply joints. A support vector machine model is trained with a leave-one-subject-out cross validation to validate the learned measures compared to traditional statistical measures. The computational results showed >70% classification accuracy on average based on the dataset of 48 subjects (25 with CAI and 23 normal controls) examined in our designed experiment. It is found that CAI can be observed from other joints (e.g., hips) significantly, which reflects the fact that there are interactions in the multi-joint coordination system. The developed method presents a potential to support the decisions with motion patterns during diagnosis, treatment, rehabilitation of gait abnormality caused by physical injury (e.g., ankle sprains in this study) or even central nervous system disorders.
A Network-based Multimodal Data Fusion Approach for Characterizing Dynamic Multimodal Physiological Patterns
Jan 03, 2019


Abstract:Characterizing the dynamic interactive patterns of complex systems helps gain in-depth understanding of how components interrelate with each other while performing certain functions as a whole. In this study, we present a novel multimodal data fusion approach to construct a complex network, which models the interactions of biological subsystems in the human body under emotional states through physiological responses. Joint recurrence plot and temporal network metrics are employed to integrate the multimodal information at the signal level. A benchmark public dataset of is used for evaluating our model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge