Christof Meile
Multimodal registration of FISH and nanoSIMS images using convolutional neural network models
Jan 14, 2022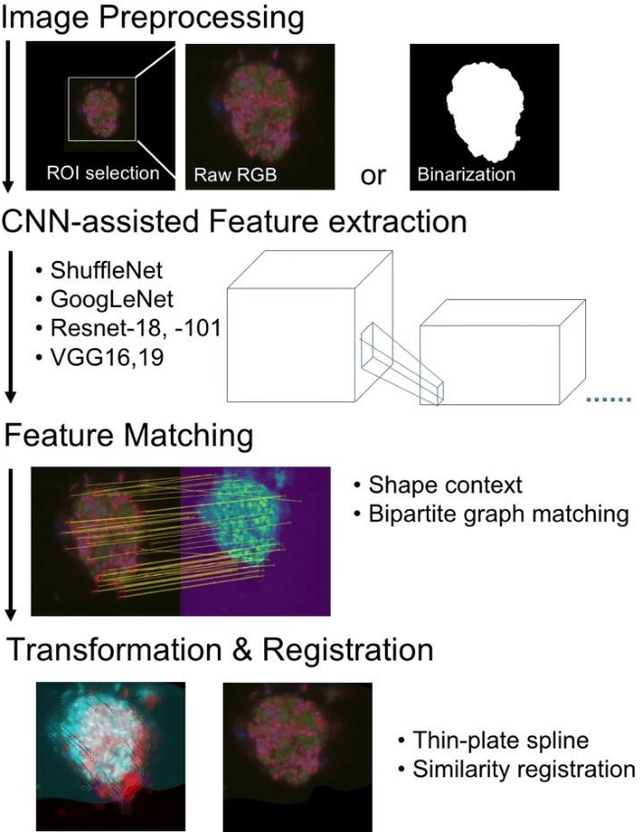
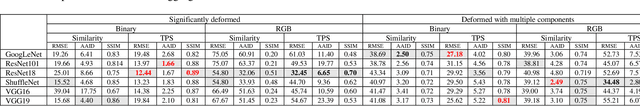
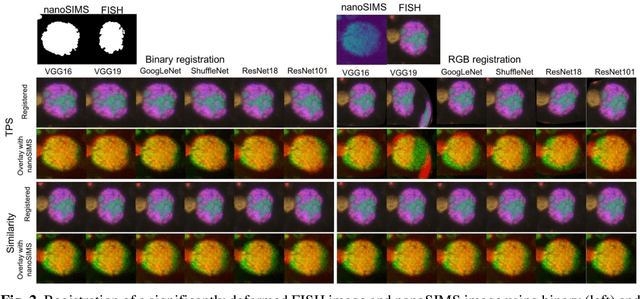
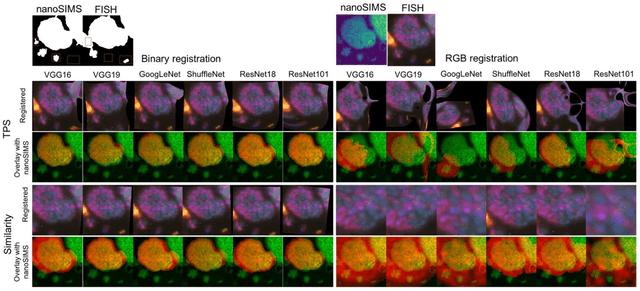
Abstract:Nanoscale secondary ion mass spectrometry (nanoSIMS) and fluorescence in situ hybridization (FISH) microscopy provide high-resolution, multimodal image representations of the identity and cell activity respectively of targeted microbial communities in microbiological research. Despite its importance to microbiologists, multimodal registration of FISH and nanoSIMS images is challenging given the morphological distortion and background noise in both images. In this study, we use convolutional neural networks (CNNs) for multiscale feature extraction, shape context for computation of the minimum transformation cost feature matching and the thin-plate spline (TPS) model for multimodal registration of the FISH and nanoSIMS images. All the six tested CNN models, VGG16, VGG19, GoogLeNet and ShuffleNet, ResNet18 and ResNet101 performed well, demonstrating the utility of CNNs in the registration of multimodal images with significant background noise and morphology distortion. We also show aggregate shape preserved by binarization to be a robust feature for registering multimodal microbiology-related images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge