Chao-Hui Huang
QuST: QuPath Extension for Integrative Whole Slide Image and Spatial Transcriptomics Analysis
May 31, 2024Abstract:Recently, various technologies have been introduced into digital pathology, including artificial intelligence (AI) driven methods, in both areas of pathological whole slide image (WSI) analysis and spatial transcriptomics (ST) analysis. AI-driven WSI analysis utilizes the power of deep learning (DL), expands the field of view for histopathological image analysis. On the other hand, ST bridges the gap between tissue spatial analysis and biological signals, offering the possibility to understand the spatial biology. However, a major bottleneck in DL-based WSI analysis is the preparation of training patterns, as hematoxylin \& eosin (H\&E) staining does not provide direct biological evidence, such as gene expression, for determining the category of a biological component. On the other hand, as of now, the resolution in ST is far beyond that of WSI, resulting the challenge of further spatial analysis. Although various WSI analysis tools, including QuPath, have cited the use of WSI analysis tools in the context of ST analysis, its usage is primarily focused on initial image analysis, with other tools being utilized for more detailed transcriptomic analysis. As a result, the information hidden beneath WSI has not yet been fully utilized to support ST analysis. To bridge this gap, we introduce QuST, a QuPath extension designed to bridge the gap between H\&E WSI and ST analyzing tasks. In this paper, we highlight the importance of integrating DL-based WSI analysis and ST analysis in understanding disease biology and the challenges in integrating these modalities due to differences in data formats and analytical methods. The QuST source code is hosted on GitHub and documentation is available at https://github.com/huangch/qust.
eXclusive Autoencoder (XAE) for Nucleus Detection and Classification on Hematoxylin and Eosin (H&E) Stained Histopathological Images
Nov 27, 2018



Abstract:In this paper, we introduced a novel feature extraction approach, named exclusive autoencoder (XAE), which is a supervised version of autoencoder (AE), able to largely improve the performance of nucleus detection and classification on hematoxylin and eosin (H&E) histopathological images. The proposed XAE can be used in any AE-based algorithm, as long as the data labels are also provided in the feature extraction phase. In the experiments, we evaluated the performance of an approach which is the combination of an XAE and a fully connected neural network (FCN) and compared with some AE-based methods. For a nucleus detection problem (considered as a nucleus/non-nucleus classification problem) on breast cancer H&E images, the F-score of the proposed XAE+FCN approach achieved 96.64% while the state-of-the-art was at 84.49%. For nucleus classification on colorectal cancer H&E images, with the annotations of four categories of epithelial, inflammatory, fibroblast and miscellaneous nuclei. The F-score of the proposed method reached 70.4%. We also proposed a lymphocyte segmentation method. In the step of lymphocyte detection, we have compared with cutting-edge technology and gained improved performance from 90% to 98.67%. We also proposed an algorithm for lymphocyte segmentation based on nucleus detection and classification. The obtained Dice coefficient achieved 88.31% while the cutting-edge approach was at 74%.
Her2 Challenge Contest: A Detailed Assessment of Automated Her2 Scoring Algorithms in Whole Slide Images of Breast Cancer Tissues
Jul 24, 2017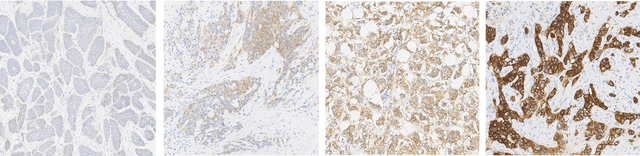
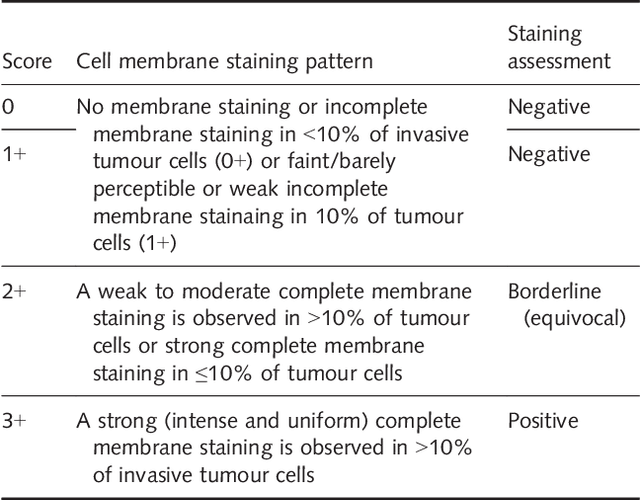
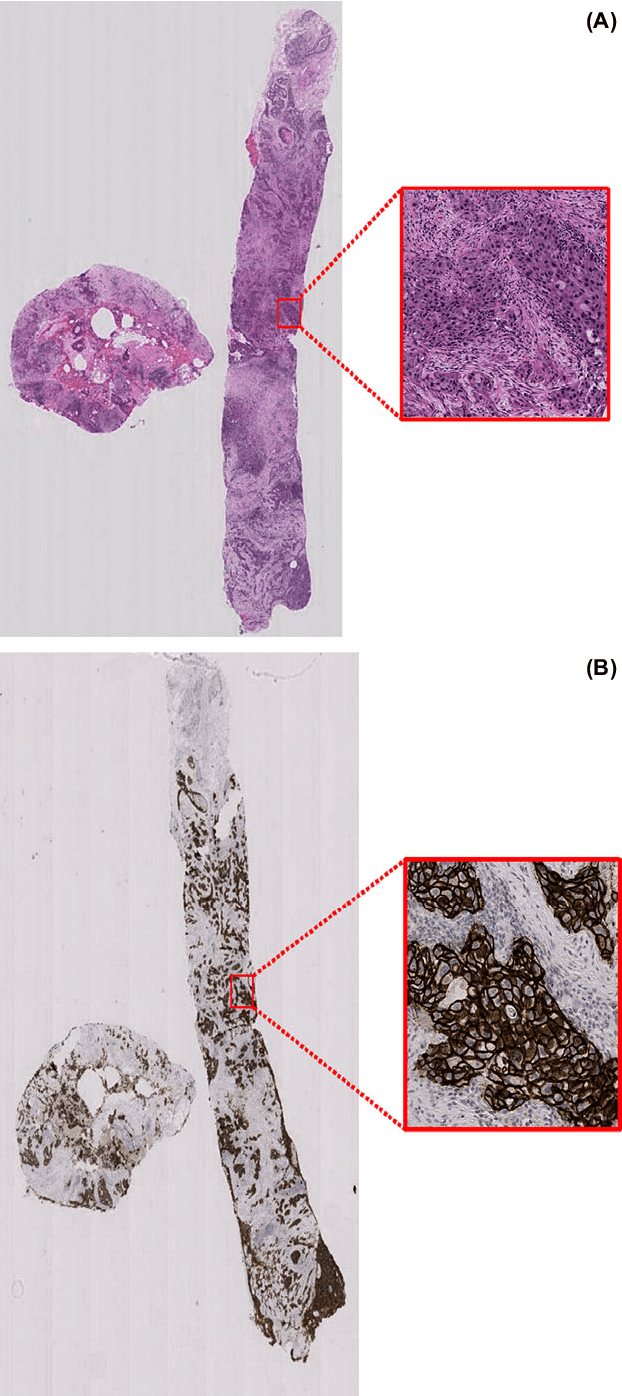
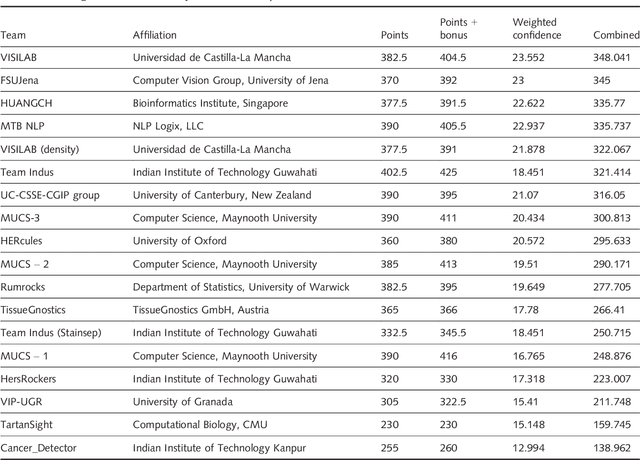
Abstract:Evaluating expression of the Human epidermal growth factor receptor 2 (Her2) by visual examination of immunohistochemistry (IHC) on invasive breast cancer (BCa) is a key part of the diagnostic assessment of BCa due to its recognised importance as a predictive and prognostic marker in clinical practice. However, visual scoring of Her2 is subjective and consequently prone to inter-observer variability. Given the prognostic and therapeutic implications of Her2 scoring, a more objective method is required. In this paper, we report on a recent automated Her2 scoring contest, held in conjunction with the annual PathSoc meeting held in Nottingham in June 2016, aimed at systematically comparing and advancing the state-of-the-art Artificial Intelligence (AI) based automated methods for Her2 scoring. The contest dataset comprised of digitised whole slide images (WSI) of sections from 86 cases of invasive breast carcinoma stained with both Haematoxylin & Eosin (H&E) and IHC for Her2. The contesting algorithms automatically predicted scores of the IHC slides for an unseen subset of the dataset and the predicted scores were compared with the 'ground truth' (a consensus score from at least two experts). We also report on a simple Man vs Machine contest for the scoring of Her2 and show that the automated methods could beat the pathology experts on this contest dataset. This paper presents a benchmark for comparing the performance of automated algorithms for scoring of Her2. It also demonstrates the enormous potential of automated algorithms in assisting the pathologist with objective IHC scoring.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge