Carlos Ortiz-de-Solórzano
NaroNet: Discovery of tumor microenvironment elements from highly multiplexed images
Mar 25, 2021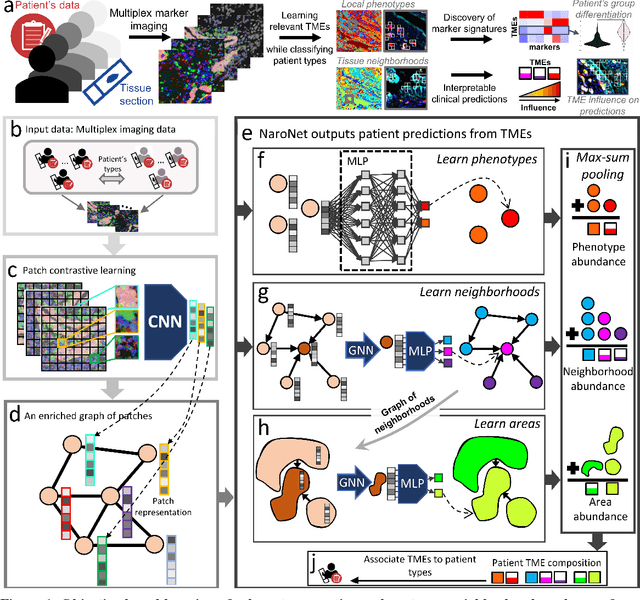
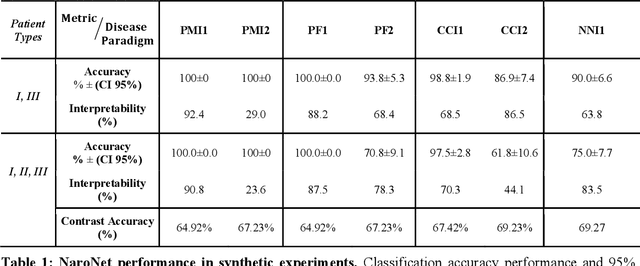
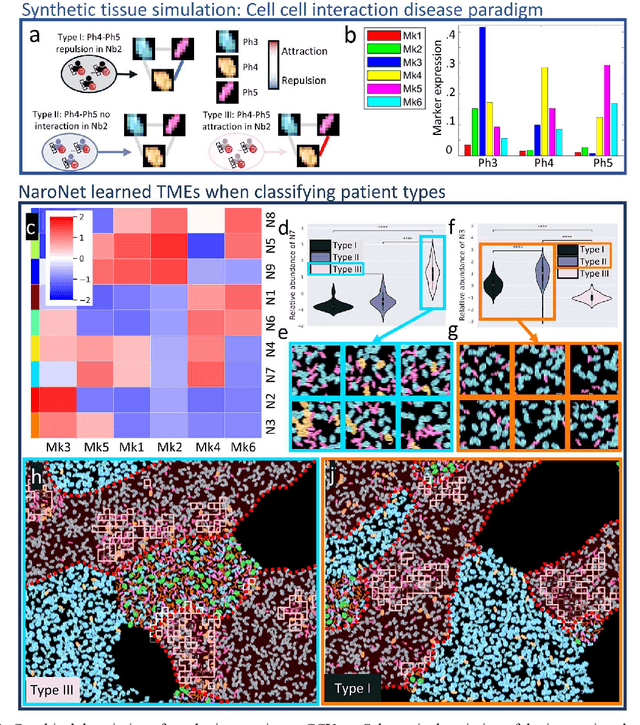
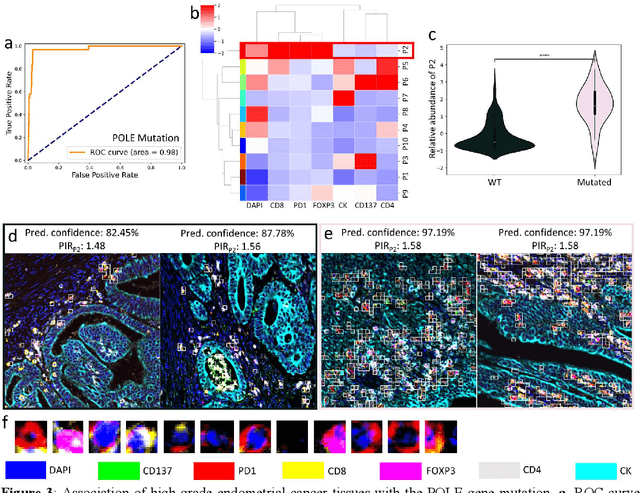
Abstract:Many efforts have been made to discover tumor-specific microenvironment elements (TMEs) from immunostained tissue sections. However, the identification of yet unknown but relevant TMEs from multiplex immunostained tissues remains a challenge, due to the number of markers involved (tens) and the complexity of their spatial interactions. We present NaroNet, which uses machine learning to identify and annotate known as well as novel TMEs from self-supervised embeddings of cells, organized at different levels (local cell phenotypes and cellular neighborhoods). Then it uses the abundance of TMEs to classify patients based on biological or clinical features. We validate NaroNet using synthetic patient cohorts with adjustable incidence of different TMEs and two cancer patient datasets. In both synthetic and real datasets, NaroNet unsupervisedly identifies novel TMEs, relevant for the user-defined classification task. As NaroNet requires only patient-level information, it renders state-of-the-art computational methods accessible to a broad audience, accelerating the discovery of biomarker signatures.
Synplex: A synthetic simulator of highly multiplexed histological images
Mar 08, 2021



Abstract:Multiplex tissue immunostaining is a technology of growing relevance as it can capture in situ the complex interactions existing between the elements of the tumor microenvironment. The existence and availability of large, annotated image datasets is key for the objective development and benchmarking of bioimage analysis algorithms. Manual annotation of multiplex images, is however, laborious, often impracticable. In this paper, we present Synplex, a simulation system able to generate multiplex immunostained in situ tissue images based on user-defined parameters. This includes the specification of structural attributes, such as the number of cell phenotypes, the number and level of expression of cellular markers, or the cell morphology. Synplex consists of three sequential modules, each being responsible for a separate task: modeling of cellular neighborhoods, modeling of cell phenotypes, and synthesis of realistic cell/tissue textures. Synplex flexibility and accuracy are demonstrated qualitatively and quantitatively by generating synthetic tissues that simulate disease paradigms found in the real scenarios. Synplex is publicly available for scientific purposes, and we believe it will become a valuable tool for the training and/or validation of multiplex image analysis algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge