NaroNet: Discovery of tumor microenvironment elements from highly multiplexed images
Paper and Code
Mar 25, 2021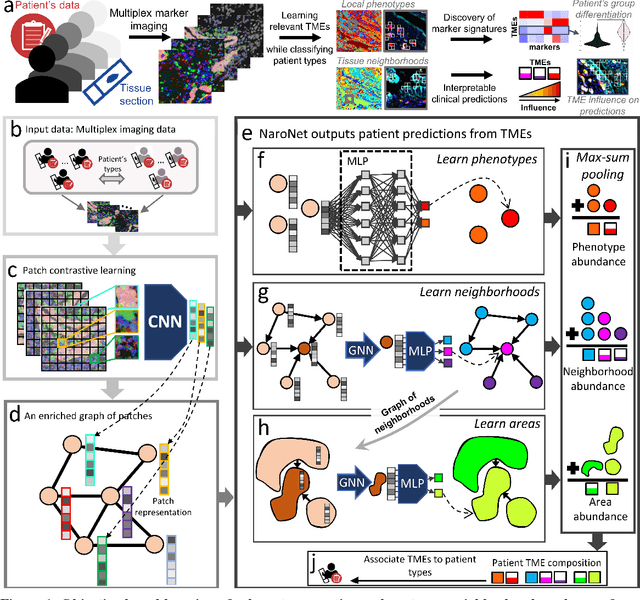
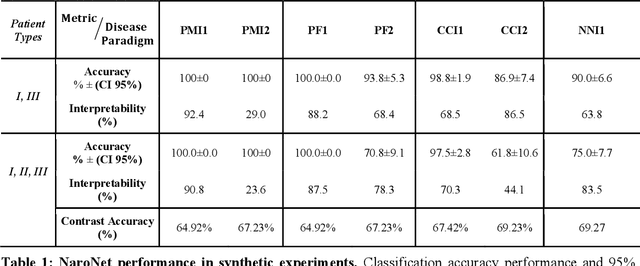
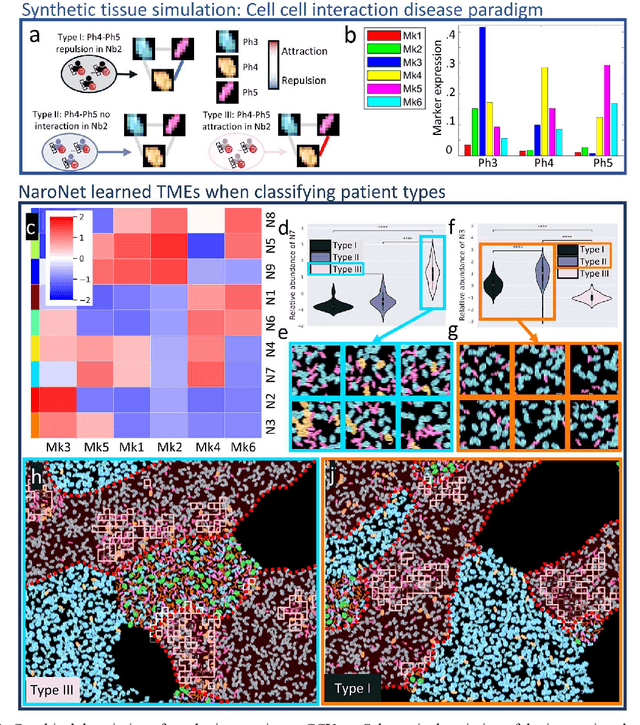
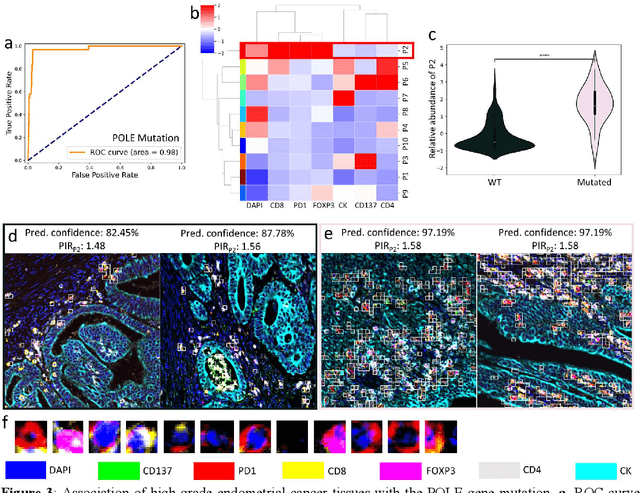
Many efforts have been made to discover tumor-specific microenvironment elements (TMEs) from immunostained tissue sections. However, the identification of yet unknown but relevant TMEs from multiplex immunostained tissues remains a challenge, due to the number of markers involved (tens) and the complexity of their spatial interactions. We present NaroNet, which uses machine learning to identify and annotate known as well as novel TMEs from self-supervised embeddings of cells, organized at different levels (local cell phenotypes and cellular neighborhoods). Then it uses the abundance of TMEs to classify patients based on biological or clinical features. We validate NaroNet using synthetic patient cohorts with adjustable incidence of different TMEs and two cancer patient datasets. In both synthetic and real datasets, NaroNet unsupervisedly identifies novel TMEs, relevant for the user-defined classification task. As NaroNet requires only patient-level information, it renders state-of-the-art computational methods accessible to a broad audience, accelerating the discovery of biomarker signatures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge