Bella Specktor-Fadida
From Cold Start to Active Learning: Embedding-Based Scan Selection for Medical Image Segmentation
Jan 26, 2026Abstract:Accurate segmentation annotations are critical for disease monitoring, yet manual labeling remains a major bottleneck due to the time and expertise required. Active learning (AL) alleviates this burden by prioritizing informative samples for annotation, typically through a diversity-based cold-start phase followed by uncertainty-driven selection. We propose a novel cold-start sampling strategy that combines foundation-model embeddings with clustering, including automatic selection of the number of clusters and proportional sampling across clusters, to construct a diverse and representative initial training. This is followed by an uncertainty-based AL framework that integrates spatial diversity to guide sample selection. The proposed method is intuitive and interpretable, enabling visualization of the feature-space distribution of candidate samples. We evaluate our approach on three datasets spanning X-ray and MRI modalities. On the CheXmask dataset, the cold-start strategy outperforms random selection, improving Dice from 0.918 to 0.929 and reducing the Hausdorff distance from 32.41 to 27.66 mm. In the AL setting, combined entropy and diversity selection improves Dice from 0.919 to 0.939 and reduces the Hausdorff distance from 30.10 to 19.16 mm. On the Montgomery dataset, cold-start gains are substantial, with Dice improving from 0.928 to 0.950 and Hausdorff distance decreasing from 14.22 to 9.38 mm. On the SynthStrip dataset, cold-start selection slightly affects Dice but reduces the Hausdorff distance from 9.43 to 8.69 mm, while active learning improves Dice from 0.816 to 0.826 and reduces the Hausdorff distance from 7.76 to 6.38 mm. Overall, the proposed framework consistently outperforms baseline methods in low-data regimes, improving segmentation accuracy.
Advances in Automated Fetal Brain MRI Segmentation and Biometry: Insights from the FeTA 2024 Challenge
May 05, 2025



Abstract:Accurate fetal brain tissue segmentation and biometric analysis are essential for studying brain development in utero. The FeTA Challenge 2024 advanced automated fetal brain MRI analysis by introducing biometry prediction as a new task alongside tissue segmentation. For the first time, our diverse multi-centric test set included data from a new low-field (0.55T) MRI dataset. Evaluation metrics were also expanded to include the topology-specific Euler characteristic difference (ED). Sixteen teams submitted segmentation methods, most of which performed consistently across both high- and low-field scans. However, longitudinal trends indicate that segmentation accuracy may be reaching a plateau, with results now approaching inter-rater variability. The ED metric uncovered topological differences that were missed by conventional metrics, while the low-field dataset achieved the highest segmentation scores, highlighting the potential of affordable imaging systems when paired with high-quality reconstruction. Seven teams participated in the biometry task, but most methods failed to outperform a simple baseline that predicted measurements based solely on gestational age, underscoring the challenge of extracting reliable biometric estimates from image data alone. Domain shift analysis identified image quality as the most significant factor affecting model generalization, with super-resolution pipelines also playing a substantial role. Other factors, such as gestational age, pathology, and acquisition site, had smaller, though still measurable, effects. Overall, FeTA 2024 offers a comprehensive benchmark for multi-class segmentation and biometry estimation in fetal brain MRI, underscoring the need for data-centric approaches, improved topological evaluation, and greater dataset diversity to enable clinically robust and generalizable AI tools.
SegQC: a segmentation network-based framework for multi-metric segmentation quality control and segmentation error detection in volumetric medical images
Nov 12, 2024Abstract:Quality control of structures segmentation in volumetric medical images is important for identifying segmentation errors in clinical practice and for facilitating model development. This paper introduces SegQC, a novel framework for segmentation quality estimation and segmentation error detection. SegQC computes an estimate measure of the quality of a segmentation in volumetric scans and in their individual slices and identifies possible segmentation error regions within a slice. The key components include: 1. SegQC-Net, a deep network that inputs a scan and its segmentation mask and outputs segmentation error probabilities for each voxel in the scan; 2. three new segmentation quality metrics, two overlap metrics and a structure size metric, computed from the segmentation error probabilities; 3. a new method for detecting possible segmentation errors in scan slices computed from the segmentation error probabilities. We introduce a new evaluation scheme to measure segmentation error discrepancies based on an expert radiologist corrections of automatically produced segmentations that yields smaller observer variability and is closer to actual segmentation errors. We demonstrate SegQC on three fetal structures in 198 fetal MRI scans: fetal brain, fetal body and the placenta. To assess the benefits of SegQC, we compare it to the unsupervised Test Time Augmentation (TTA)-based quality estimation. Our studies indicate that SegQC outperforms TTA-based quality estimation in terms of Pearson correlation and MAE for fetal body and fetal brain structures segmentation. Our segmentation error detection method achieved recall and precision rates of 0.77 and 0.48 for fetal body, and 0.74 and 0.55 for fetal brain segmentation error detection respectively. SegQC enhances segmentation metrics estimation for whole scans and individual slices, as well as provides error regions detection.
Test-time augmentation-based active learning and self-training for label-efficient segmentation
Aug 21, 2023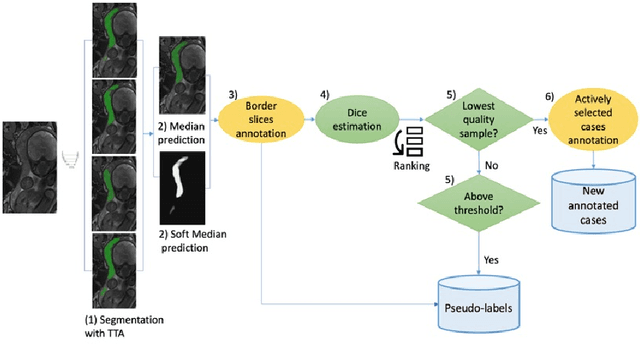
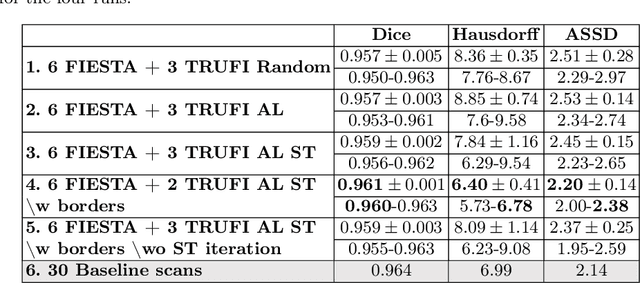
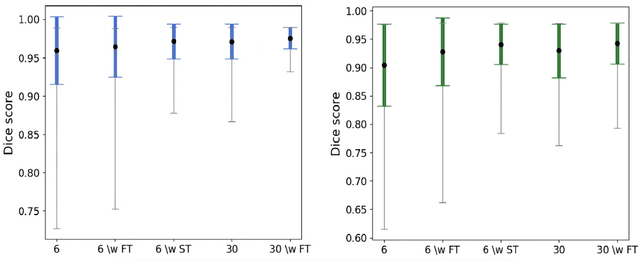
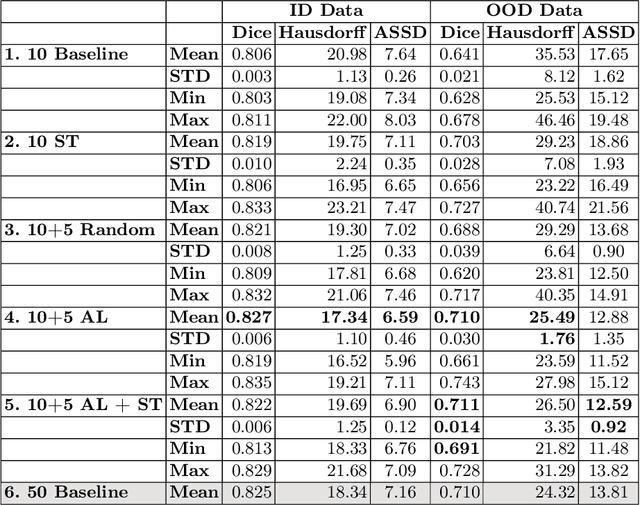
Abstract:Deep learning techniques depend on large datasets whose annotation is time-consuming. To reduce annotation burden, the self-training (ST) and active-learning (AL) methods have been developed as well as methods that combine them in an iterative fashion. However, it remains unclear when each method is the most useful, and when it is advantageous to combine them. In this paper, we propose a new method that combines ST with AL using Test-Time Augmentations (TTA). First, TTA is performed on an initial teacher network. Then, cases for annotation are selected based on the lowest estimated Dice score. Cases with high estimated scores are used as soft pseudo-labels for ST. The selected annotated cases are trained with existing annotated cases and ST cases with border slices annotations. We demonstrate the method on MRI fetal body and placenta segmentation tasks with different data variability characteristics. Our results indicate that ST is highly effective for both tasks, boosting performance for in-distribution (ID) and out-of-distribution (OOD) data. However, while self-training improved the performance of single-sequence fetal body segmentation when combined with AL, it slightly deteriorated performance of multi-sequence placenta segmentation on ID data. AL was helpful for the high variability placenta data, but did not improve upon random selection for the single-sequence body data. For fetal body segmentation sequence transfer, combining AL with ST following ST iteration yielded a Dice of 0.961 with only 6 original scans and 2 new sequence scans. Results using only 15 high-variability placenta cases were similar to those using 50 cases. Code is available at: https://github.com/Bella31/TTA-quality-estimation-ST-AL
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge