Artur Luczak
Reinforcement Learning with Brain-Inspired Modulation can Improve Adaptation to Environmental Changes
May 19, 2022



Abstract:Developments in reinforcement learning (RL) have allowed algorithms to achieve impressive performance in highly complex, but largely static problems. In contrast, biological learning seems to value efficiency of adaptation to a constantly-changing world. Here we build on a recently-proposed neuronal learning rule that assumes each neuron can optimize its energy balance by predicting its own future activity. That assumption leads to a neuronal learning rule that uses presynaptic input to modulate prediction error. We argue that an analogous RL rule would use action probability to modulate reward prediction error. This modulation makes the agent more sensitive to negative experiences, and more careful in forming preferences. We embed the proposed rule in both tabular and deep-Q-network RL algorithms, and find that it outperforms conventional algorithms in simple, but highly-dynamic tasks. We suggest that the new rule encapsulates a core principle of biological intelligence; an important component for allowing algorithms to adapt to change in a human-like way.
Hippocluster: an efficient, hippocampus-inspired algorithm for graph clustering
May 19, 2022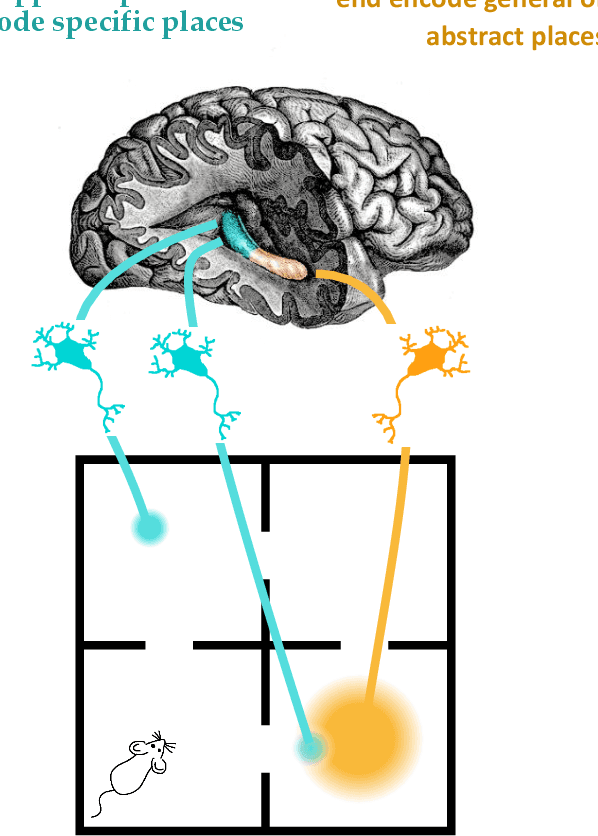
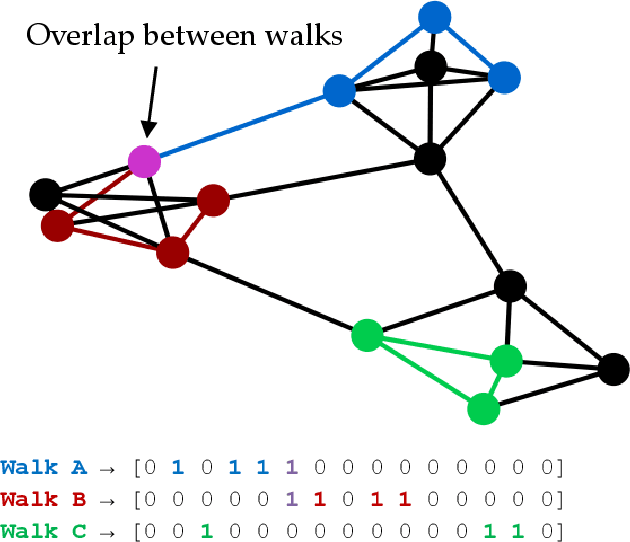
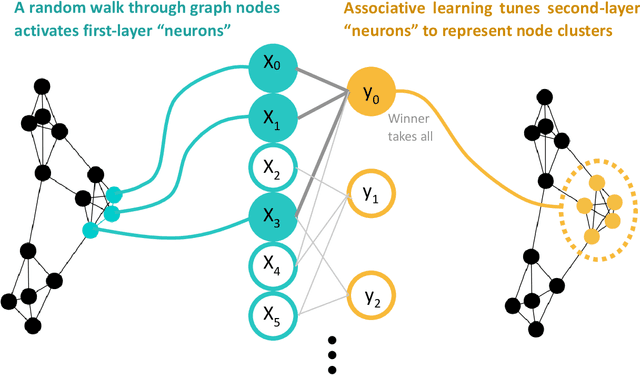

Abstract:Random walks can reveal communities or clusters in networks, because they are more likely to stay within a cluster than leave it. Thus, one family of community detection algorithms uses random walks to measure distance between pairs of nodes in various ways, and then applies K-Means or other generic clustering methods to these distances. Interestingly, information processing in the brain may suggest a simpler method of learning clusters directly from random walks. Drawing inspiration from the hippocampus, we describe a simple two-layer neural learning framework. Neurons in one layer are associated with graph nodes and simulate random walks. These simulations cause neurons in the second layer to become tuned to graph clusters through simple associative learning. We show that if these neuronal interactions are modelled a particular way, the system is essentially a variant of K-Means clustering applied directly in the walk-space, bypassing the usual step of computing node distances/similarities. The result is an efficient graph clustering method. Biological information processing systems are known for high efficiency and adaptability. In tests on benchmark graphs, our framework demonstrates this high data-efficiency, low memory use, low complexity, and real-time adaptation to graph changes, while still achieving clustering quality comparable to other algorithms.
A Deep Convolutional Auto-Encoder with Pooling - Unpooling Layers in Caffe
Jan 18, 2017



Abstract:This paper presents the development of several models of a deep convolutional auto-encoder in the Caffe deep learning framework and their experimental evaluation on the example of MNIST dataset. We have created five models of a convolutional auto-encoder which differ architecturally by the presence or absence of pooling and unpooling layers in the auto-encoder's encoder and decoder parts. Our results show that the developed models provide very good results in dimensionality reduction and unsupervised clustering tasks, and small classification errors when we used the learned internal code as an input of a supervised linear classifier and multi-layer perceptron. The best results were provided by a model where the encoder part contains convolutional and pooling layers, followed by an analogous decoder part with deconvolution and unpooling layers without the use of switch variables in the decoder part. The paper also discusses practical details of the creation of a deep convolutional auto-encoder in the very popular Caffe deep learning framework. We believe that our approach and results presented in this paper could help other researchers to build efficient deep neural network architectures in the future.
Creation of a Deep Convolutional Auto-Encoder in Caffe
Apr 22, 2016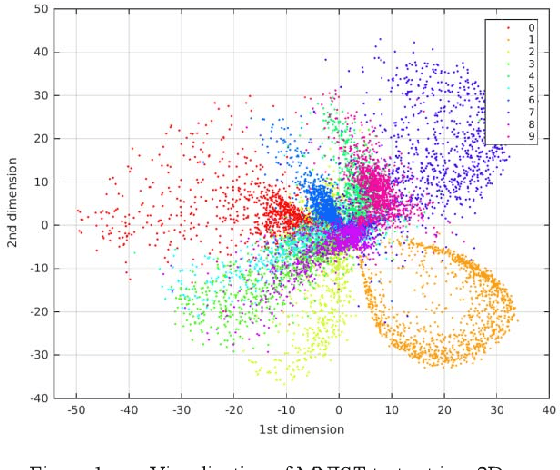
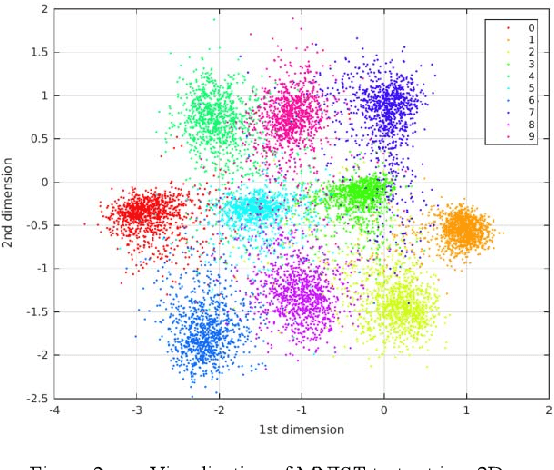
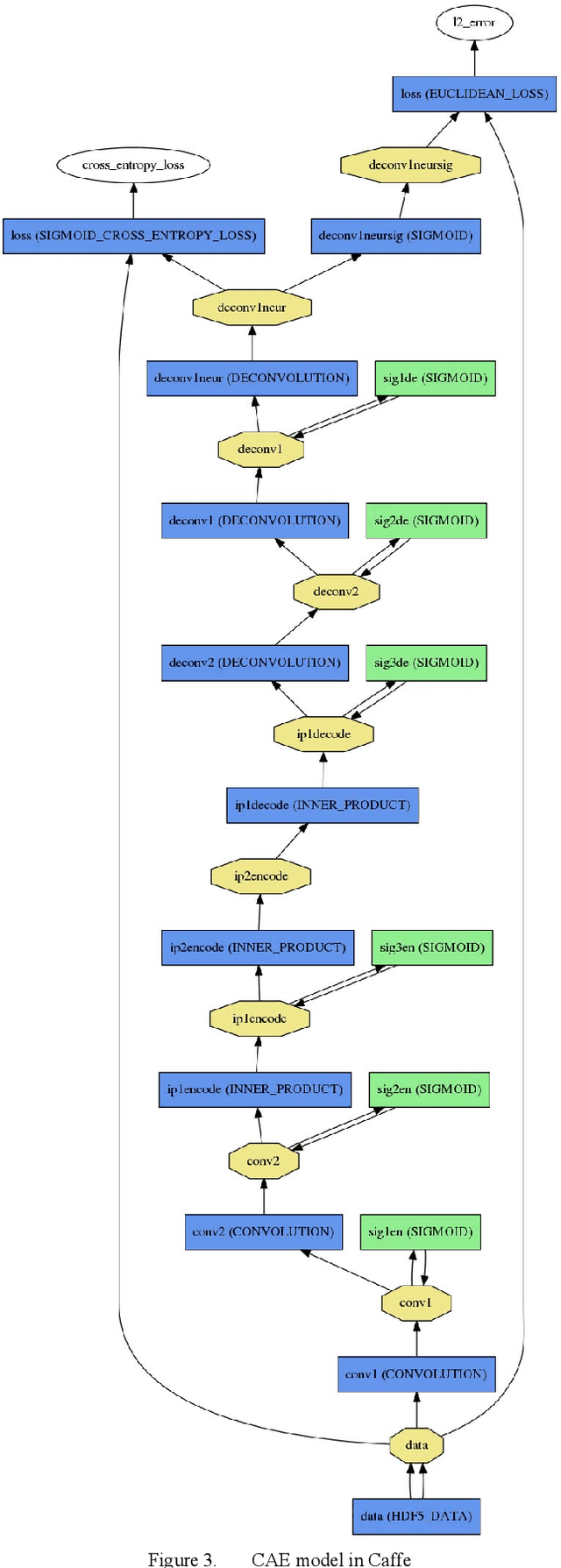
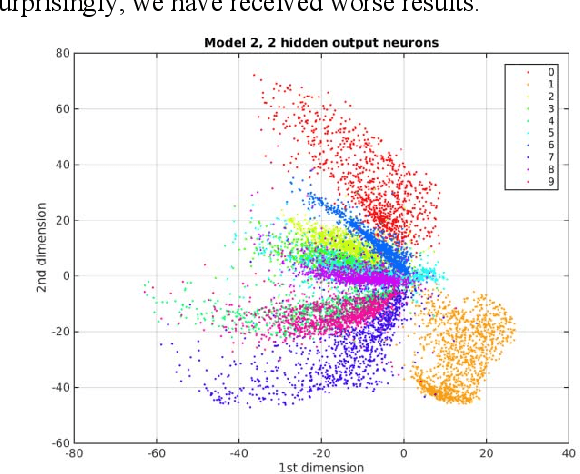
Abstract:The development of a deep (stacked) convolutional auto-encoder in the Caffe deep learning framework is presented in this paper. We describe simple principles which we used to create this model in Caffe. The proposed model of convolutional auto-encoder does not have pooling/unpooling layers yet. The results of our experimental research show comparable accuracy of dimensionality reduction in comparison with a classic auto-encoder on the example of MNIST dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge