Arthur Imbert
PointFISH -- learning point cloud representations for RNA localization patterns
Feb 21, 2023Abstract:Subcellular RNA localization is a critical mechanism for the spatial control of gene expression. Its mechanism and precise functional role is not yet very well understood. Single Molecule Fluorescence in Situ Hybridization (smFISH) images allow for the detection of individual RNA molecules with subcellular accuracy. In return, smFISH requires robust methods to quantify and classify RNA spatial distribution. Here, we present PointFISH, a novel computational approach for the recognition of RNA localization patterns. PointFISH is an attention-based network for computing continuous vector representations of RNA point clouds. Trained on simulations only, it can directly process extracted coordinates from experimental smFISH images. The resulting embedding allows scalable and flexible spatial transcriptomics analysis and matches performance of hand-crafted pipelines.
Learning with minimal effort: leveraging in silico labeling for cell and nucleus segmentation
Jan 10, 2023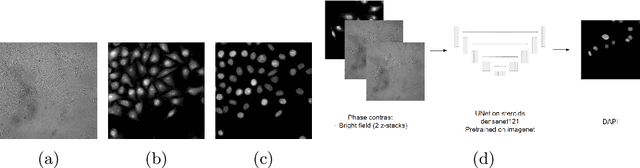
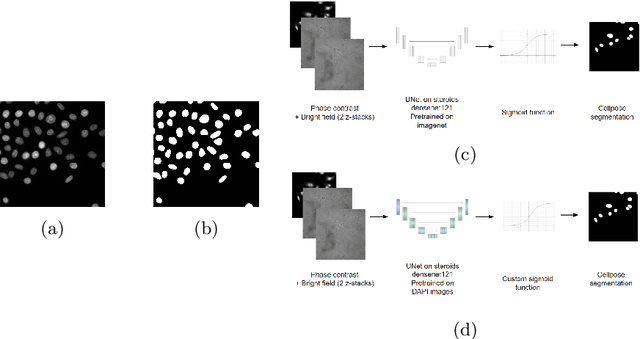
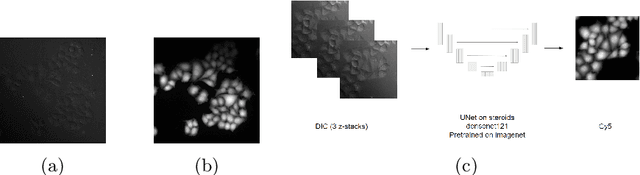
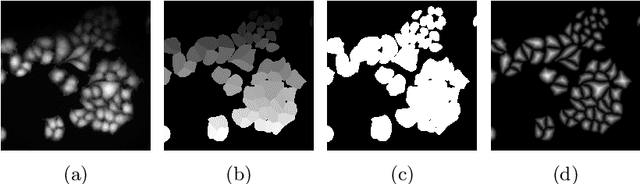
Abstract:Deep learning provides us with powerful methods to perform nucleus or cell segmentation with unprecedented quality. However, these methods usually require large training sets of manually annotated images, which are tedious and expensive to generate. In this paper we propose to use In Silico Labeling (ISL) as a pretraining scheme for segmentation tasks. The strategy is to acquire label-free microscopy images (such as bright-field or phase contrast) along fluorescently labeled images (such as DAPI or CellMask). We then train a model to predict the fluorescently labeled images from the label-free microscopy images. By comparing segmentation performance across several training set sizes, we show that such a scheme can dramatically reduce the number of required annotations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge