Antonello Vidiri
Deep learning can differentiate IDH-mutant from IDH-wild type GBM
Feb 24, 2021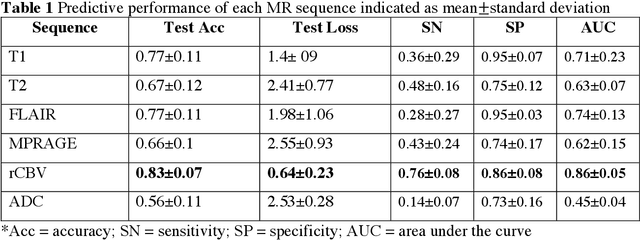
Abstract:Background: Distinction of IDH mutant and wildtype GBMs is challenging on MRI, since conventional imaging shows considerable overlap. While few studies employed deep-learning in a mixed low/high grade glioma population, a GBM-specific model is still lacking in the literature. Our objective was to develop a deep-learning model for IDH prediction in GBM by using Convoluted Neural Networks (CNN) on multiparametric MRI. Methods: We included 100 adult patients with pathologically proven GBM and IDH testing. MRI data included: morphologic sequences, rCBV and ADC maps. Tumor area was obtained by a bounding box function on the axial slice with widest tumor extension on T2 images and was projected on every sequence. Data was split into training and test (80:20) sets. A 4 block 2D - CNN architecture was implemented for IDH prediction on every MRI sequence. IDH mutation probability was calculated with softmax activation function from the last dense layer. Highest performance was calculated accounting for model accuracy and categorical cross-entropy loss (CCEL) in the test cohort. Results: Our model achieved the following performance: T1 (accuracy 77%, CCEL 1.4), T2 (accuracy 67%, CCEL 2.41), FLAIR (accuracy 77%, CCEL 1.98), MPRAGE (accuracy 66%, CCEL 2.55), rCBV (accuracy 83%, CCEL 0.64). ADC achieved lower performance. Conclusion: We built a GBM-tailored deep-learning model for IDH mutation prediction, achieving accuracy of 83% with rCBV maps. High predictivity of perfusion images may reflect the known correlation between IDH, hypoxia inducible factor (HIF) and neoangiogenesis. This model may set a path for non-invasive evaluation of IDH mutation in GBM.
Comparison of Machine Learning Classifiers to Predict Patient Survival and Genetics of GBM: Towards a Standardized Model for Clinical Implementation
Feb 10, 2021



Abstract:Radiomic models have been shown to outperform clinical data for outcome prediction in glioblastoma (GBM). However, clinical implementation is limited by lack of parameters standardization. We aimed to compare nine machine learning classifiers, with different optimization parameters, to predict overall survival (OS), isocitrate dehydrogenase (IDH) mutation, O-6-methylguanine-DNA-methyltransferase (MGMT) promoter methylation, epidermal growth factor receptor (EGFR) VII amplification and Ki-67 expression in GBM patients, based on radiomic features from conventional and advanced MR. 156 adult patients with pathologic diagnosis of GBM were included. Three tumoral regions were analyzed: contrast-enhancing tumor, necrosis and non-enhancing tumor, selected by manual segmentation. Radiomic features were extracted with a custom version of Pyradiomics, and selected through Boruta algorithm. A Grid Search algorithm was applied when computing 4 times K-fold cross validation (K=10) to get the highest mean and lowest spread of accuracy. Once optimal parameters were identified, model performances were assessed in terms of Area Under The Curve-Receiver Operating Characteristics (AUC-ROC). Metaheuristic and ensemble classifiers showed the best performance across tasks. xGB obtained maximum accuracy for OS (74.5%), AB for IDH mutation (88%), MGMT methylation (71,7%), Ki-67 expression (86,6%), and EGFR amplification (81,6%). Best performing features shed light on possible correlations between MR and tumor histology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge