Anna Palczewska
Identification of pattern mining algorithm for rugby league players positional groups separation based on movement patterns
Feb 25, 2023



Abstract:The application of pattern mining algorithms to extract movement patterns from sports big data can improve training specificity by facilitating a more granular evaluation of movement. As there are various pattern mining algorithms, this study aimed to validate which algorithm discovers the best set of movement patterns for player movement profiling in professional rugby league and the similarity in extracted movement patterns between the algorithms. Three pattern mining algorithms (l-length Closed Contiguous [LCCspm], Longest Common Subsequence [LCS] and AprioriClose) were used to profile elite rugby football league hookers (n = 22 players) and wingers (n = 28 players) match-games movements across 319 matches. Machine learning classification algorithms were used to identify which algorithm gives the best set of movement patterns to separate playing positions with Jaccard similarity score identifying the extent of similarity between algorithms' movement patterns. LCCspm and LCS movement patterns shared a 0.19 Jaccard similarity score. AprioriClose movement patterns shared no significant similarity with LCCspm and LCS patterns. The closed contiguous movement patterns profiled by LCCspm best-separated players into playing positions. Multi-layered Perceptron algorithm achieved the highest accuracy of 91.02% and precision, recall and F1 scores of 0.91 respectively. Therefore, we recommend the extraction of closed contiguous (consecutive) over non-consecutive movement patterns for separating groups of players.
Development of an expected possession value model to analyse team attacking performances in rugby league
May 05, 2021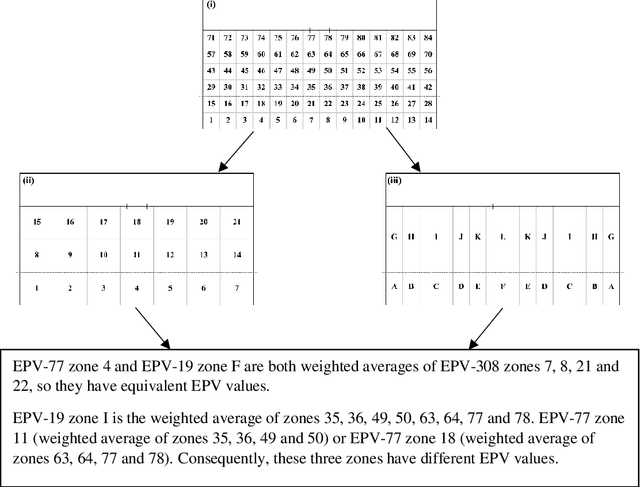
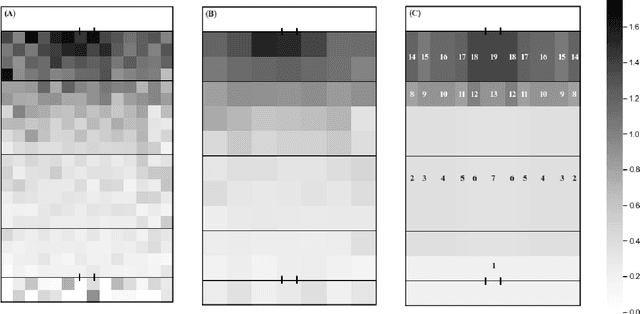
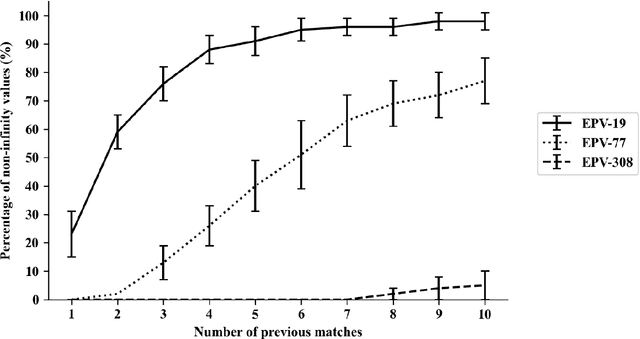
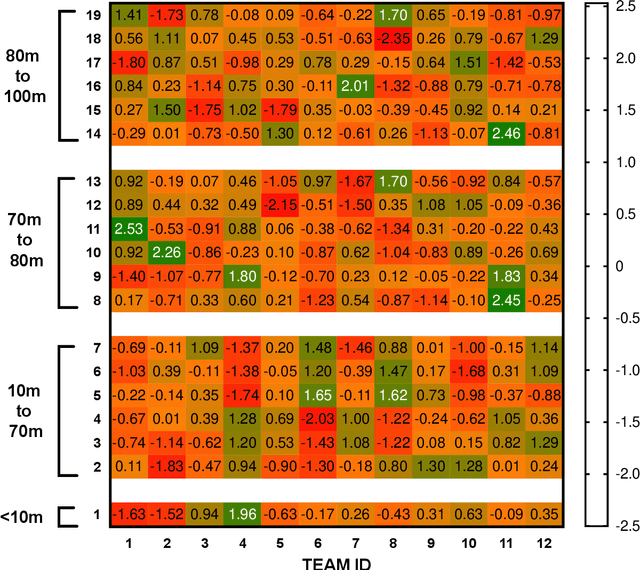
Abstract:This study aimed to provide a framework to evaluate team attacking performances in rugby league using 59,233 plays from 180 Super League matches via expected possession value (EPV) models. The EPV-308 split the pitch into 308 5m x 5m zones, the EPV-77 split the pitch into 77 10m x 10m zones and the EPV-19 split the pitch in 19 zones of variable size dependent on the total zone value generated during a match. Attacking possessions were considered as Markov Chains, allowing the value of each zone visited to be estimated based on the outcome of the possession. The Kullback-Leibler Divergence was used to evaluate the reproducibility of the value generated from each zone (the reward distribution) by teams between matches. The EPV-308 had the greatest variability and lowest reproducibility, compared to EPV-77 and EPV-19. When six previous matches were considered, the team's subsequent match attacking performances had a similar reward distribution for EPV-19, EPV-77 and EPV-308 on 95 +/- 4%, 51 +/- 12% and 0 +/- 0% of occasions. This study supports the use of EPV-19 to evaluate team attacking performance in rugby league and provides a simple framework through which attacking performances can be compared between teams.
Interpreting random forest classification models using a feature contribution method
Dec 04, 2013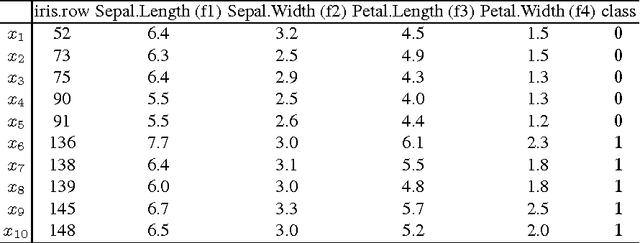
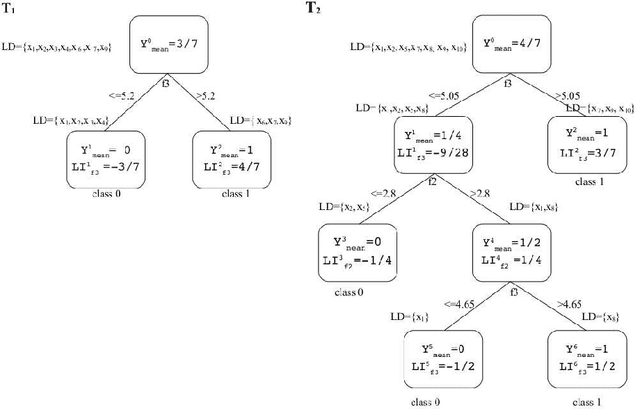
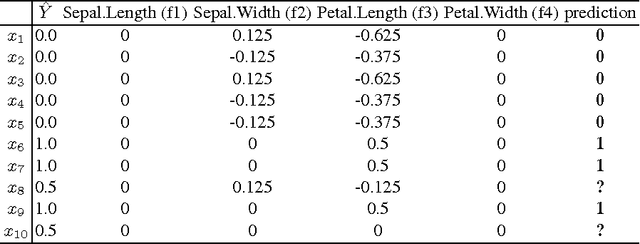
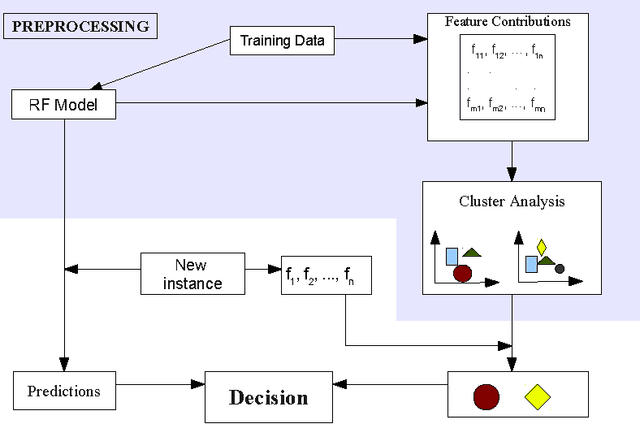
Abstract:Model interpretation is one of the key aspects of the model evaluation process. The explanation of the relationship between model variables and outputs is relatively easy for statistical models, such as linear regressions, thanks to the availability of model parameters and their statistical significance. For "black box" models, such as random forest, this information is hidden inside the model structure. This work presents an approach for computing feature contributions for random forest classification models. It allows for the determination of the influence of each variable on the model prediction for an individual instance. By analysing feature contributions for a training dataset, the most significant variables can be determined and their typical contribution towards predictions made for individual classes, i.e., class-specific feature contribution "patterns", are discovered. These patterns represent a standard behaviour of the model and allow for an additional assessment of the model reliability for a new data. Interpretation of feature contributions for two UCI benchmark datasets shows the potential of the proposed methodology. The robustness of results is demonstrated through an extensive analysis of feature contributions calculated for a large number of generated random forest models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge