Alina Enescu
Improved Slice-wise Tumour Detection in Brain MRIs by Computing Dissimilarities between Latent Representations
Jul 24, 2020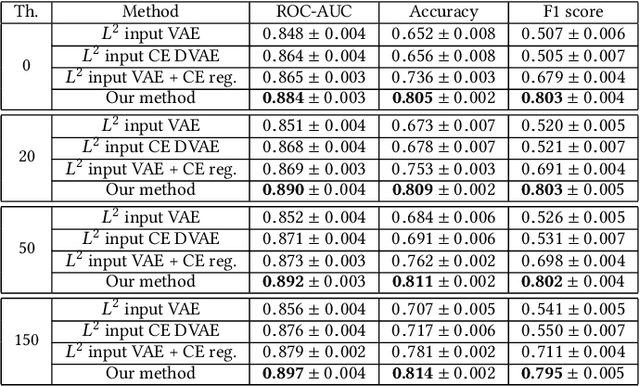
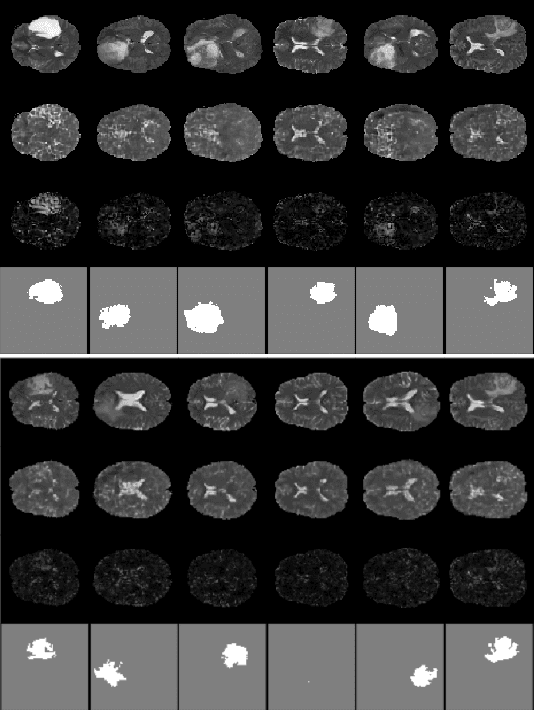
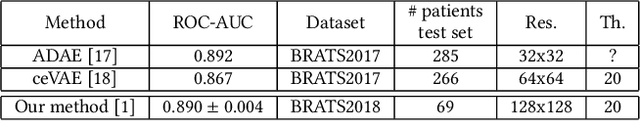
Abstract:Anomaly detection for Magnetic Resonance Images (MRIs) can be solved with unsupervised methods by learning the distribution of healthy images and identifying anomalies as outliers. In presence of an additional dataset of unlabelled data containing also anomalies, the task can be framed as a semi-supervised task with negative and unlabelled sample points. Recently, in Albu et al., 2020, we have proposed a slice-wise semi-supervised method for tumour detection based on the computation of a dissimilarity function in the latent space of a Variational AutoEncoder, trained on unlabelled data. The dissimilarity is computed between the encoding of the image and the encoding of its reconstruction obtained through a different autoencoder trained only on healthy images. In this paper we present novel and improved results for our method, obtained by training the Variational AutoEncoders on a subset of the HCP and BRATS-2018 datasets and testing on the remaining individuals. We show that by training the models on higher resolution images and by improving the quality of the reconstructions, we obtain results which are comparable with different baselines, which employ a single VAE trained on healthy individuals. As expected, the performance of our method increases with the size of the threshold used to determine the presence of an anomaly.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge