Ali Mohammadi Shanghooshabad
Graphical Join: A New Physical Join Algorithm for RDBMSs
Jun 22, 2022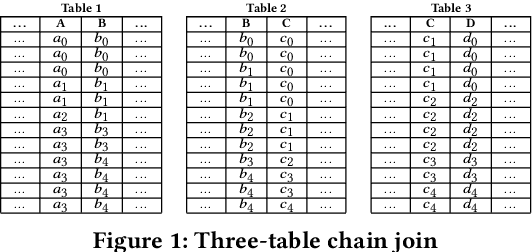
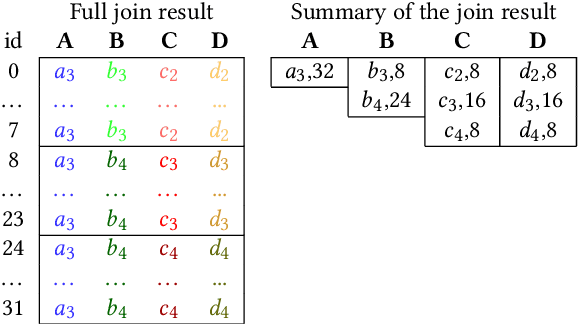
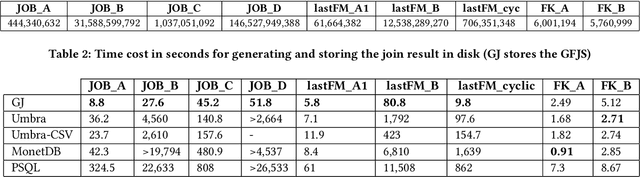
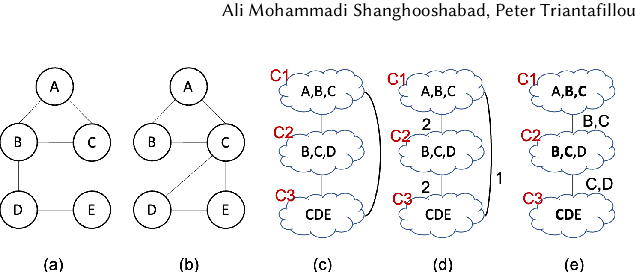
Abstract:Join operations (especially n-way, many-to-many joins) are known to be time- and resource-consuming. At large scales, with respect to table and join-result sizes, current state of the art approaches (including both binary-join plans which use Nested-loop/Hash/Sort-merge Join algorithms or, alternatively, worst-case optimal join algorithms (WOJAs)), may even fail to produce any answer given reasonable resource and time constraints. In this work, we introduce a new approach for n-way equi-join processing, the Graphical Join (GJ). The key idea is two-fold: First, to map the physical join computation problem to PGMs and introduce tweaked inference algorithms which can compute a Run-Length Encoding (RLE) based join-result summary, entailing all statistics necessary to materialize the join result. Second, and most importantly, to show that a join algorithm, like GJ, which produces the above join-result summary and then desummarizes it, can introduce large performance benefits in time and space. Comprehensive experimentation is undertaken with join queries from the JOB, TPCDS, and lastFM datasets, comparing GJ against PostgresQL and MonetDB and a state of the art WOJA implemented within the Umbra system. The results for in-memory join computation show performance improvements up to 64X, 388X, and 6X faster than PostgreSQL, MonetDB and Umbra, respectively. For on-disk join computation, GJ is faster than PostgreSQL, MonetDB and Umbra by up to 820X, 717X and 165X, respectively. Furthermore, GJ space needs are up to 21,488X, 38,333X, and 78,750X smaller than PostgresQL, MonetDB, and Umbra, respectively.
Model Joins: Enabling Analytics Over Joins of Absent Big Tables
Jun 21, 2022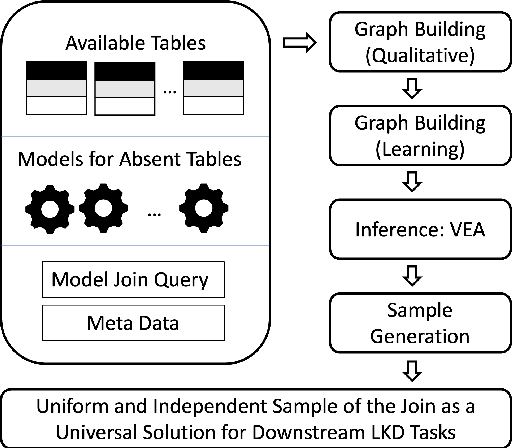
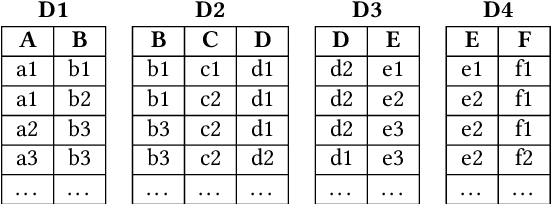
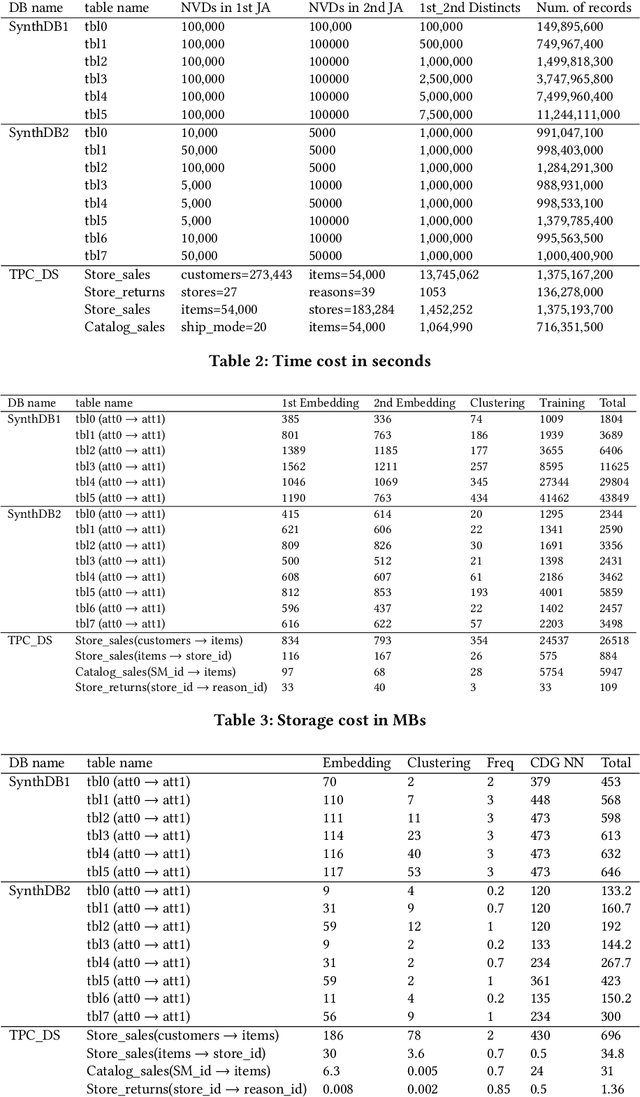
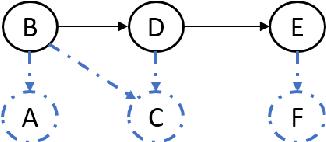
Abstract:This work is motivated by two key facts. First, it is highly desirable to be able to learn and perform knowledge discovery and analytics (LKD) tasks without the need to access raw-data tables. This may be due to organizations finding it increasingly frustrating and costly to manage and maintain ever-growing tables, or for privacy reasons. Hence, compact models can be developed from the raw data and used instead of the tables. Second, oftentimes, LKD tasks are to be performed on a (potentially very large) table which is itself the result of joining separate (potentially very large) relational tables. But how can one do this, when the individual to-be-joined tables are absent? Here, we pose the following fundamental questions: Q1: How can one "join models" of (absent/deleted) tables or "join models with other tables" in a way that enables LKD as if it were performed on the join of the actual raw tables? Q2: What are appropriate models to use per table? Q3: As the model join would be an approximation of the actual data join, how can one evaluate the quality of the model join result? This work puts forth a framework, Model Join, addressing these challenges. The framework integrates and joins the per-table models of the absent tables and generates a uniform and independent sample that is a high-quality approximation of a uniform and independent sample of the actual raw-data join. The approximation stems from the models, but not from the Model Join framework. The sample obtained by the Model Join can be used to perform LKD downstream tasks, such as approximate query processing, classification, clustering, regression, association rule mining, visualization, and so on. To our knowledge, this is the first work with this agenda and solutions. Detailed experiments with TPC-DS data and synthetic data showcase Model Join's usefulness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge