Alexandra Jankowski
Scalable Gaussian Process Regression for Kernels with a Non-Stationary Phase
Dec 25, 2019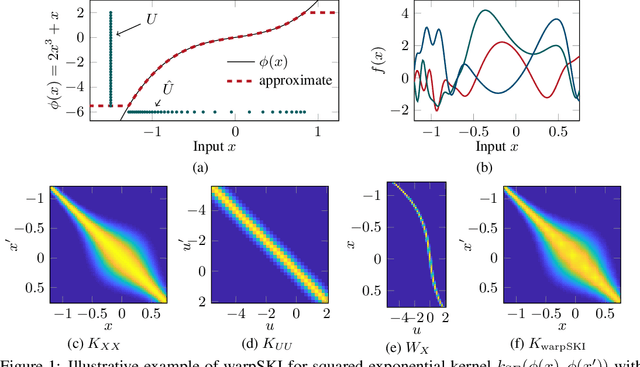
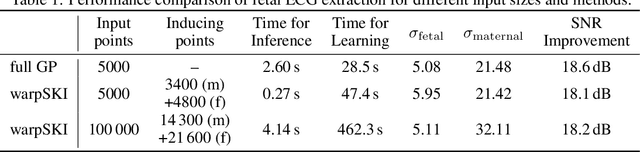
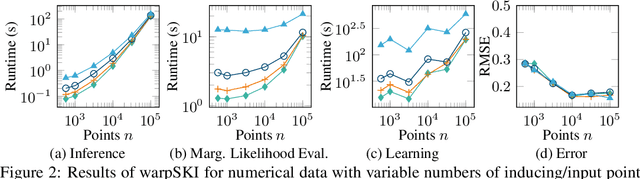

Abstract:The application of Gaussian processes (GPs) to large data sets is limited due to heavy memory and computational requirements. A variety of methods has been proposed to enable scalability, one of which is to exploit structure in the kernel matrix. Previous methods, however, cannot easily deal with non-stationary processes. This paper presents an efficient GP framework, that extends structured kernel interpolation methods to GPs with a non-stationary phase. We particularly treat mixtures of non-stationary processes, which are commonly used in the context of separation problems e.g. in biomedical signal processing. Our approach employs multiple sets of non-equidistant inducing points to account for the non-stationarity and retrieve Toeplitz and Kronecker structure in the kernel matrix allowing for efficient inference. Kernel learning is done by optimizing the marginal likelihood, which can be approximated efficiently using stochastic trace estimation methods. Our approach is demonstrated on numerical examples and large biomedical datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge