Alain Barrat
An embedding-based distance for temporal graphs
Jan 23, 2024Abstract:We define a distance between temporal graphs based on graph embeddings built using time-respecting random walks. We study both the case of matched graphs, when there exists a known relation between the nodes, and the unmatched case, when such a relation is unavailable and the graphs may be of different sizes. We illustrate the interest of our distance definition, using both real and synthetic temporal network data, by showing its ability to discriminate between graphs with different structural and temporal properties. Leveraging state-of-the-art machine learning techniques, we propose an efficient implementation of distance computation that is viable for large-scale temporal graphs.
DyANE: Dynamics-aware node embedding for temporal networks
Sep 12, 2019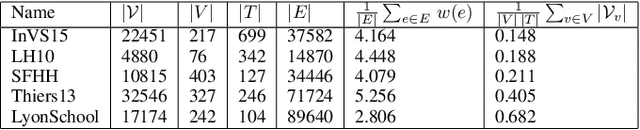
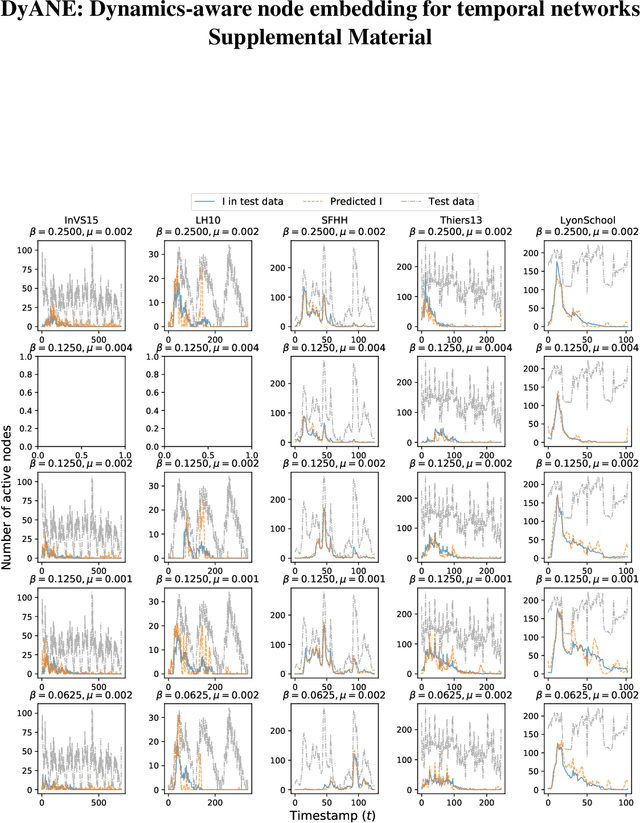
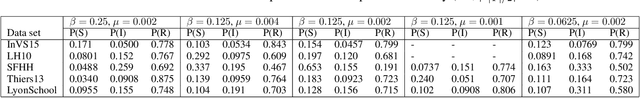
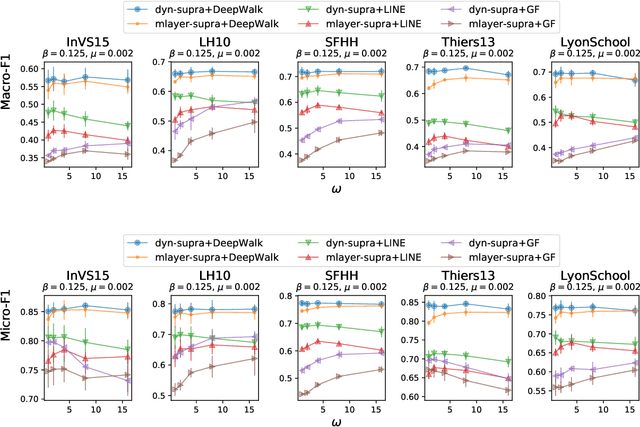
Abstract:Low-dimensional vector representations of network nodes have proven successful to feed graph data to machine learning algorithms and to improve performance across diverse tasks. Most of the embedding techniques, however, have been developed with the goal of achieving dense, low-dimensional encoding of network structure and patterns. Here, we present a node embedding technique aimed at providing low-dimensional feature vectors that are informative of dynamical processes occurring over temporal networks - rather than of the network structure itself - with the goal of enabling prediction tasks related to the evolution and outcome of these processes. We achieve this by using a modified supra-adjacency representation of temporal networks and building on standard embedding techniques for static graphs based on random-walks. We show that the resulting embedding vectors are useful for prediction tasks related to paradigmatic dynamical processes, namely epidemic spreading over empirical temporal networks. In particular, we illustrate the performance of our approach for the prediction of nodes' epidemic states in a single instance of the spreading process. We show how framing this task as a supervised multi-label classification task on the embedding vectors allows us to estimate the temporal evolution of the entire system from a partial sampling of nodes at random times, with potential impact for nowcasting infectious disease dynamics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge