A. P. Vinod
Subject Specific Deep Learning Model for Motor Imagery Direction Decoding
Jan 03, 2025



Abstract:Hemispheric strokes impair motor control in contralateral body parts, necessitating effective rehabilitation strategies. Motor Imagery-based Brain-Computer Interfaces (MI-BCIs) promote neuroplasticity, aiding the recovery of motor functions. While deep learning has shown promise in decoding MI actions for stroke rehabilitation, existing studies largely focus on bilateral MI actions and are limited to offline evaluations. Decoding directional information from unilateral MI, however, offers a more natural control interface with greater degrees of freedom but remains challenging due to spatially overlapping neural activity. This work proposes a novel deep learning framework for online decoding of binary directional MI signals from the dominant hand of 20 healthy subjects. The proposed method employs EEGNet-based convolutional filters to extract temporal and spatial features. The EEGNet model is enhanced by Squeeze-and-Excitation (SE) layers that rank the electrode importance and feature maps. A subject-independent model is initially trained using calibration data from multiple subjects and fine-tuned for subject-specific adaptation. The performance of the proposed method is evaluated using subject-specific online session data. The proposed method achieved an average right vs left binary direction decoding accuracy of 58.7 +\- 8% for unilateral MI tasks, outperforming the existing deep learning models. Additionally, the SE-layer ranking offers insights into electrode contribution, enabling potential subject-specific BCI optimization. The findings highlight the efficacy of the proposed method in advancing MI-BCI applications for a more natural and effective control of BCI systems.
FBCNet: A Multi-view Convolutional Neural Network for Brain-Computer Interface
Mar 17, 2021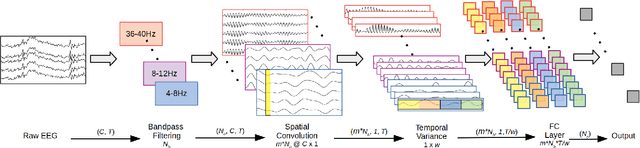
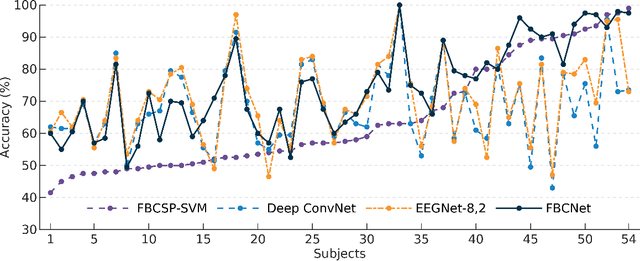
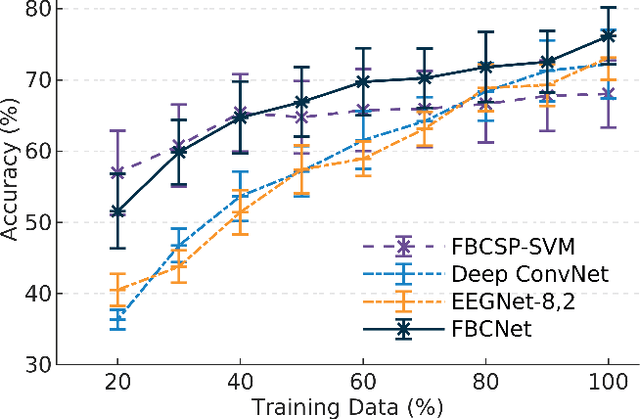
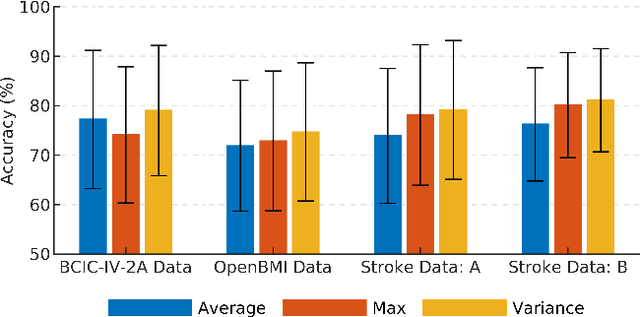
Abstract:Lack of adequate training samples and noisy high-dimensional features are key challenges faced by Motor Imagery (MI) decoding algorithms for electroencephalogram (EEG) based Brain-Computer Interface (BCI). To address these challenges, inspired from neuro-physiological signatures of MI, this paper proposes a novel Filter-Bank Convolutional Network (FBCNet) for MI classification. FBCNet employs a multi-view data representation followed by spatial filtering to extract spectro-spatially discriminative features. This multistage approach enables efficient training of the network even when limited training data is available. More significantly, in FBCNet, we propose a novel Variance layer that effectively aggregates the EEG time-domain information. With this design, we compare FBCNet with state-of-the-art (SOTA) BCI algorithm on four MI datasets: The BCI competition IV dataset 2a (BCIC-IV-2a), the OpenBMI dataset, and two large datasets from chronic stroke patients. The results show that, by achieving 76.20% 4-class classification accuracy, FBCNet sets a new SOTA for BCIC-IV-2a dataset. On the other three datasets, FBCNet yields up to 8% higher binary classification accuracies. Additionally, using explainable AI techniques we present one of the first reports about the differences in discriminative EEG features between healthy subjects and stroke patients. Also, the FBCNet source code is available at https://github.com/ravikiran-mane/FBCNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge