Vessel-CAPTCHA: an efficient learning framework for vessel annotation and segmentation
Paper and Code
Jan 29, 2021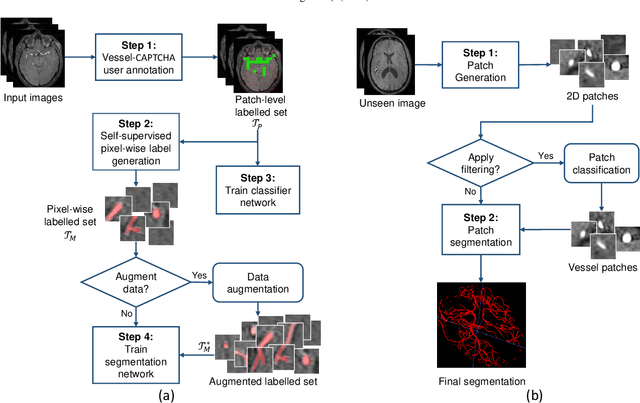
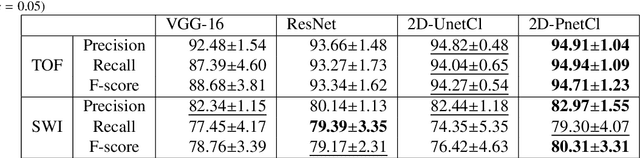
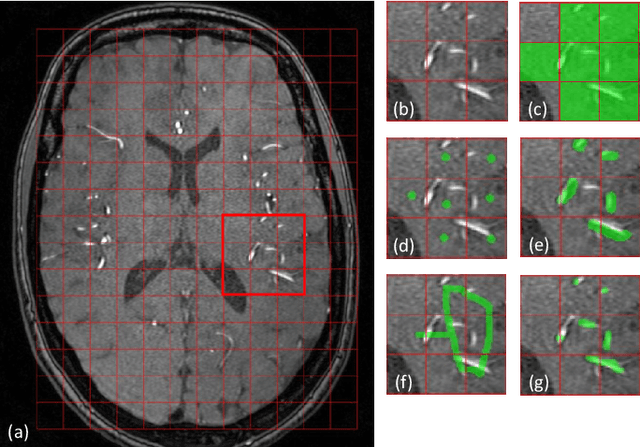
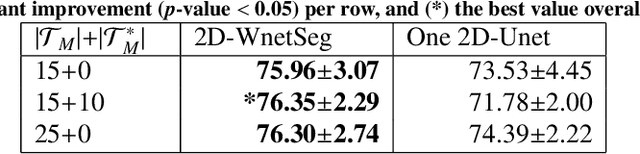
The use of deep learning techniques for 3D brain vessel image segmentation has not been as widespread as for the segmentation of other organs and tissues. This can be explained by two factors. First, deep learning techniques tend to show poor performances at the segmentation of relatively small objects compared to the size of the full image. Second, due to the complexity of vascular trees and the small size of vessels, it is challenging to obtain the amount of annotated training data typically needed by deep learning methods. To address these problems, we propose a novel annotation-efficient deep learning vessel segmentation framework. The framework avoids pixel-wise annotations, only requiring patch-level labels to discriminate between vessel and non-vessel 2D patches in the training set, in a setup similar to the CAPTCHAs used to differentiate humans from bots in web applications. The user-provided annotations are used for two tasks: 1) to automatically generate pixel-wise labels for vessels and background in each patch, which are used to train a segmentation network, and 2) to train a classifier network. The classifier network allows to generate additional weak patch labels, further reducing the annotation burden, and it acts as a noise filter for poor quality images. We use this framework for the segmentation of the cerebrovascular tree in Time-of-Flight angiography (TOF) and Susceptibility-Weighted Images (SWI). The results show that the framework achieves state-of-the-art accuracy, while reducing the annotation time by up to 80% with respect to learning-based segmentation methods using pixel-wise labels for training
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge