Using a modified double deep image prior for crosstalk mitigation in multislice ptychography
Paper and Code
Jan 29, 2021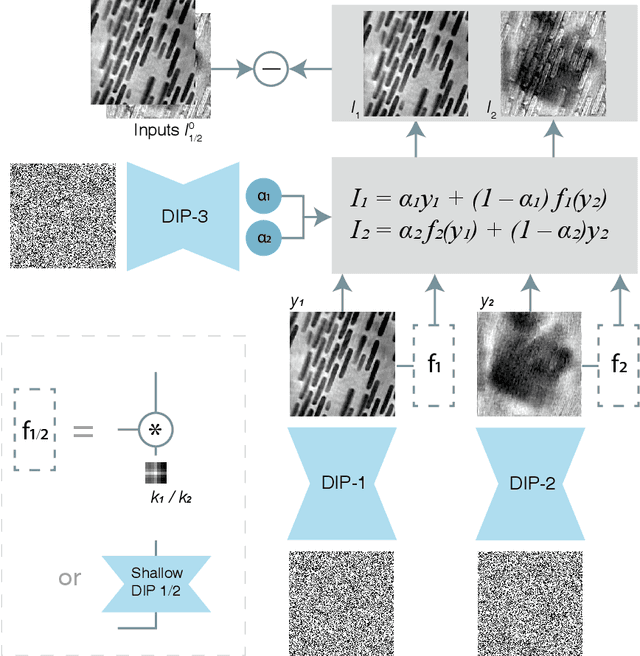
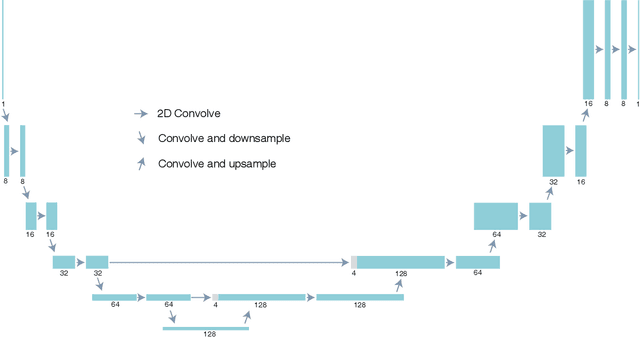
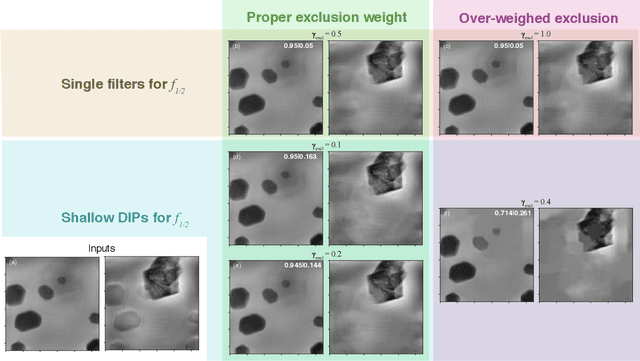
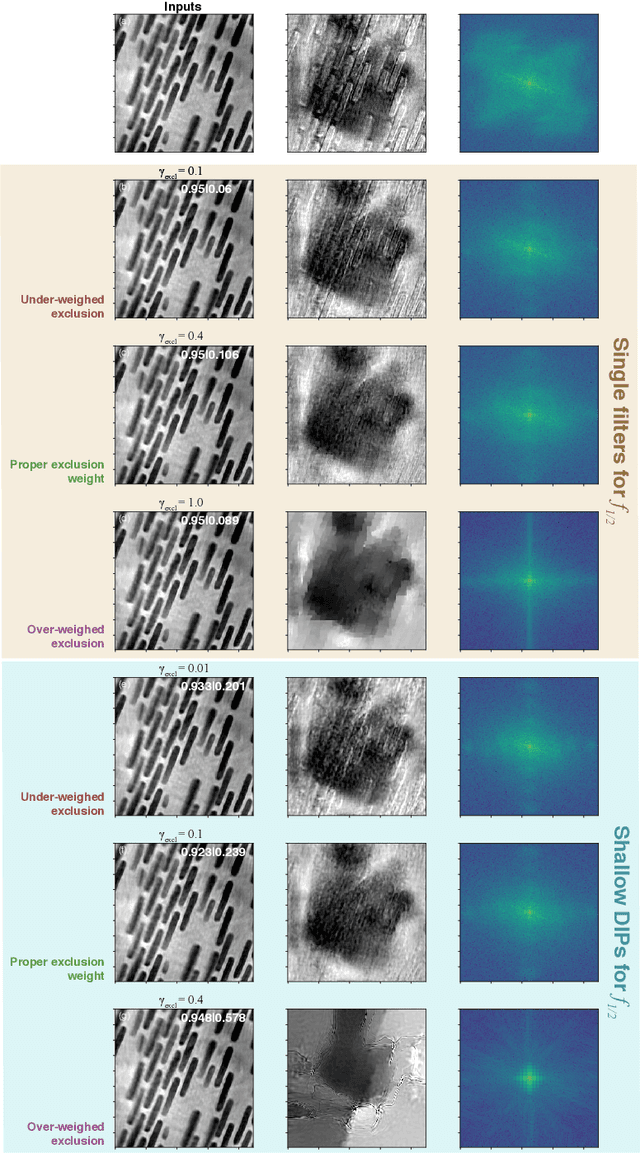
Multislice ptychography is a high-resolution microscopy technique used to image multiple separate axial planes using a single illumination direction. However, multislice ptychography reconstructions are often degraded by crosstalk, where some features on one plane erroneously contribute to the reconstructed image of another plane. Here, we demonstrate the use of a modified "double deep image prior" (DDIP) architecture in mitigating crosstalk artifacts in multislice ptychography. Utilizing the tendency of generative neural networks to produce natural images, a modified DDIP method yielded good results on experimental data. For one of the datasets, we show that using DDIP could remove the need of using additional experimental data, such as from x-ray fluorescence, to suppress the crosstalk. Our method may help x-ray multislice ptychography work for more general experimental scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge