Towards Complex Artificial Life
Paper and Code
May 16, 2018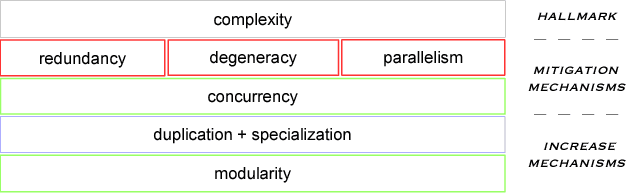
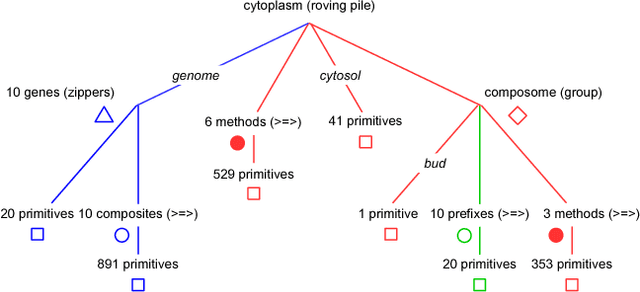
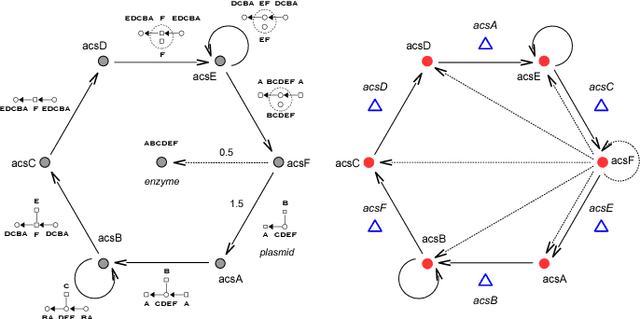
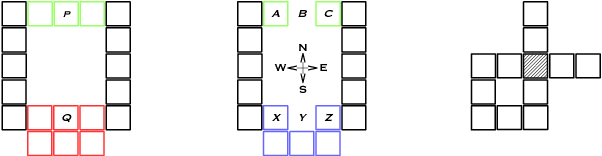
An object-oriented combinator chemistry was used to construct an artificial organism with a system architecture possessing characteristics necessary for organisms to evolve into more complex forms. This architecture supports modularity by providing a mechanism for the construction of executable modules called $methods$ that can be duplicated and specialized to increase complexity. At the same time, its support for concurrency provides the flexibility in execution order necessary for redundancy, degeneracy and parallelism to mitigate increased replication costs. The organism is a moving, self-replicating, spatially distributed assembly of elemental combinators called a $roving \: pile.$ The pile hosts an asynchronous message passing computation implemented by parallel subprocesses encoded by genes distributed through out the pile like the plasmids of a bacterial cell.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge