TopER: Topological Embeddings in Graph Representation Learning
Paper and Code
Oct 02, 2024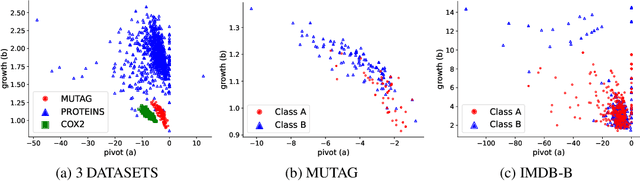
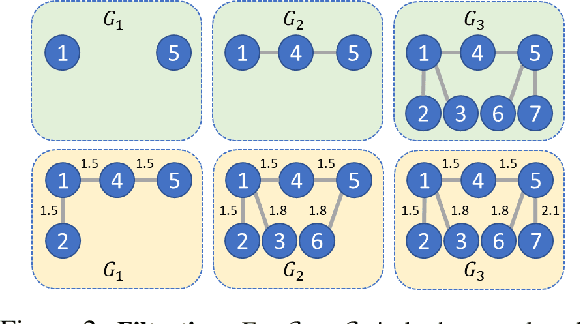
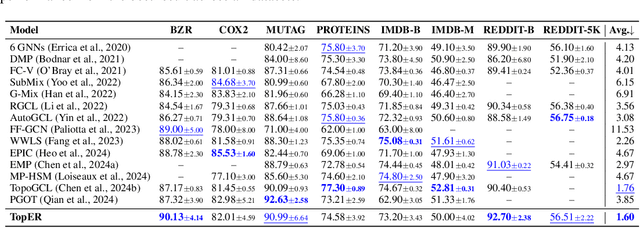
Graph embeddings play a critical role in graph representation learning, allowing machine learning models to explore and interpret graph-structured data. However, existing methods often rely on opaque, high-dimensional embeddings, limiting interpretability and practical visualization. In this work, we introduce Topological Evolution Rate (TopER), a novel, low-dimensional embedding approach grounded in topological data analysis. TopER simplifies a key topological approach, Persistent Homology, by calculating the evolution rate of graph substructures, resulting in intuitive and interpretable visualizations of graph data. This approach not only enhances the exploration of graph datasets but also delivers competitive performance in graph clustering and classification tasks. Our TopER-based models achieve or surpass state-of-the-art results across molecular, biological, and social network datasets in tasks such as classification, clustering, and visualization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge