Three principles of data science: predictability, computability, and stability (PCS)
Paper and Code
Jan 23, 2019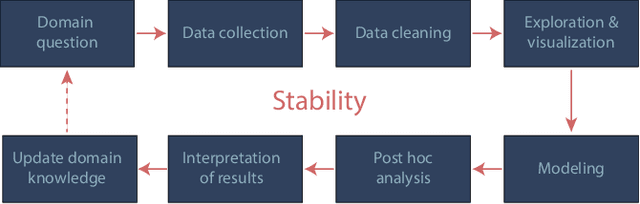
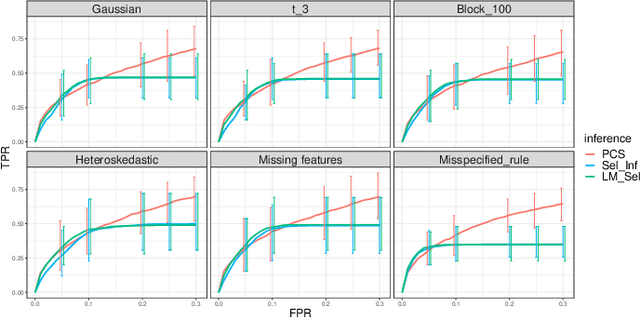
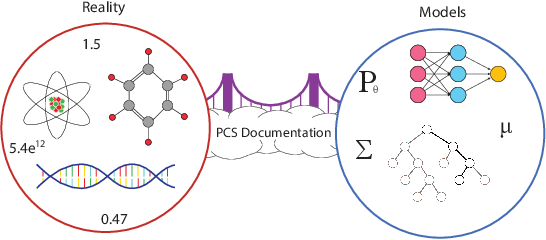
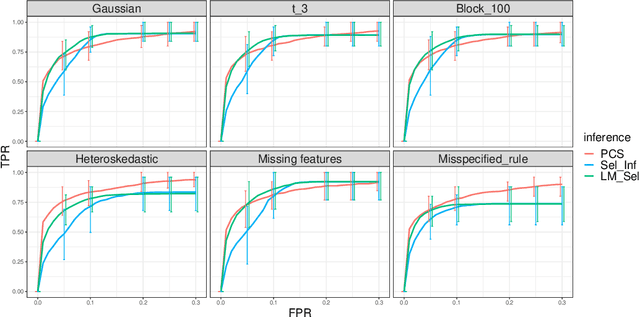
We propose the predictability, computability, and stability (PCS) framework to extract reproducible knowledge from data that can guide scientific hypothesis generation and experimental design. The PCS framework builds on key ideas in machine learning, using predictability as a reality check and evaluating computational considerations in data collection, data storage, and algorithm design. It augments PC with an overarching stability principle, which largely expands traditional statistical uncertainty considerations. In particular, stability assesses how results vary with respect to choices (or perturbations) made across the data science life cycle, including problem formulation, pre-processing, modeling (data and algorithm perturbations), and exploratory data analysis (EDA) before and after modeling. Furthermore, we develop PCS inference to investigate the stability of data results and identify when models are consistent with relatively simple phenomena. We compare PCS inference with existing methods, such as selective inference, in high-dimensional sparse linear model simulations to demonstrate that our methods consistently outperform others in terms of ROC curves over a wide range of simulation settings. Finally, we propose a PCS documentation based on Rmarkdown, iPython, or Jupyter Notebook, with publicly available, reproducible codes and narratives to back up human choices made throughout an analysis. The PCS workflow and documentation are demonstrated in a genomics case study available on Zenodo.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge