Spectral Inference Methods on Sparse Graphs: Theory and Applications
Paper and Code
Oct 14, 2016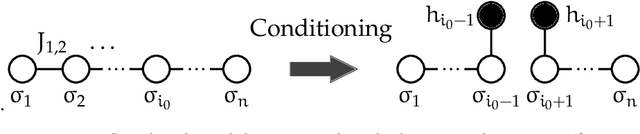
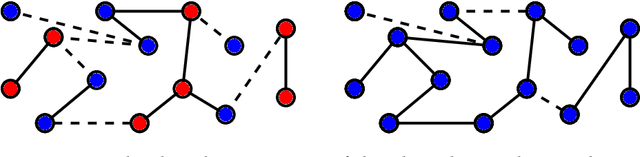
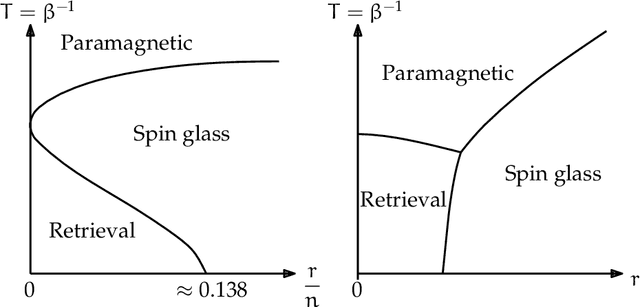
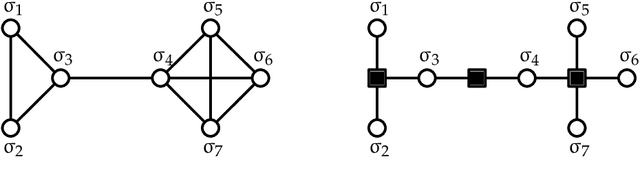
In an era of unprecedented deluge of (mostly unstructured) data, graphs are proving more and more useful, across the sciences, as a flexible abstraction to capture complex relationships between complex objects. One of the main challenges arising in the study of such networks is the inference of macroscopic, large-scale properties affecting a large number of objects, based solely on the microscopic interactions between their elementary constituents. Statistical physics, precisely created to recover the macroscopic laws of thermodynamics from an idealized model of interacting particles, provides significant insight to tackle such complex networks. In this dissertation, we use methods derived from the statistical physics of disordered systems to design and study new algorithms for inference on graphs. Our focus is on spectral methods, based on certain eigenvectors of carefully chosen matrices, and sparse graphs, containing only a small amount of information. We develop an original theory of spectral inference based on a relaxation of various mean-field free energy optimizations. Our approach is therefore fully probabilistic, and contrasts with more traditional motivations based on the optimization of a cost function. We illustrate the efficiency of our approach on various problems, including community detection, randomized similarity-based clustering, and matrix completion.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge