Sparse Meets Dense: A Hybrid Approach to Enhance Scientific Document Retrieval
Paper and Code
Jan 08, 2024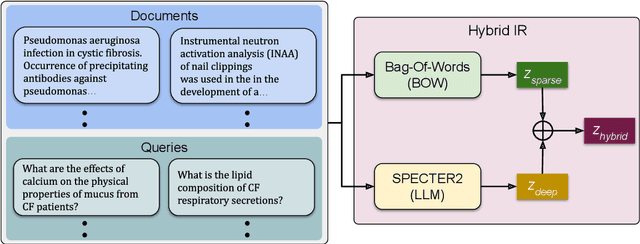
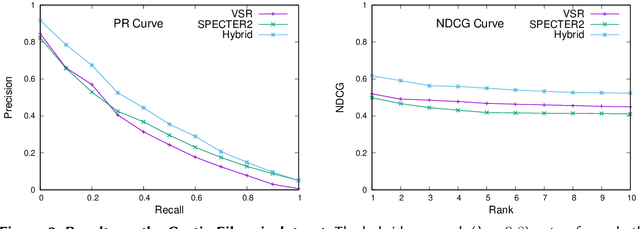
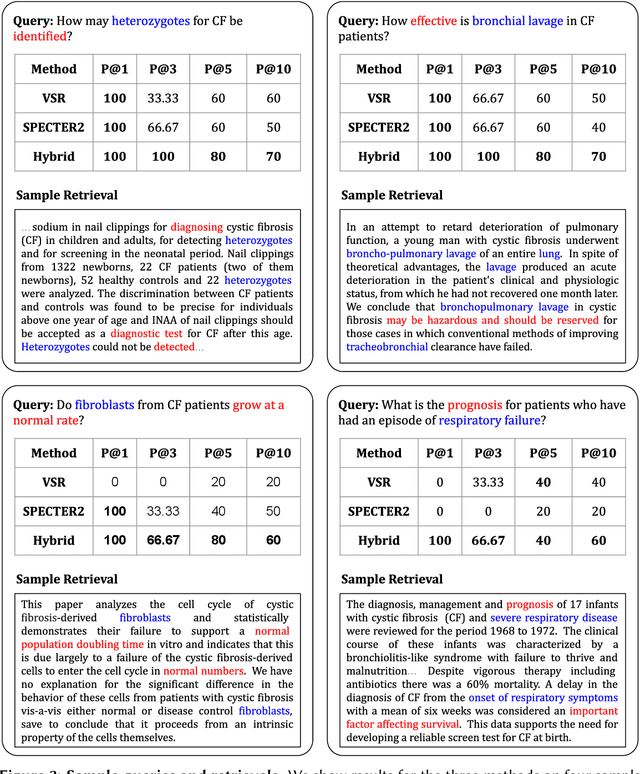
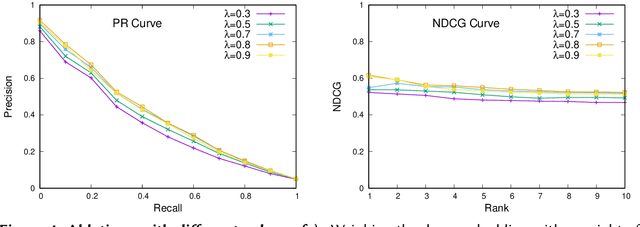
Traditional information retrieval is based on sparse bag-of-words vector representations of documents and queries. More recent deep-learning approaches have used dense embeddings learned using a transformer-based large language model. We show that on a classic benchmark on scientific document retrieval in the medical domain of cystic fibrosis, that both of these models perform roughly equivalently. Notably, dense vectors from the state-of-the-art SPECTER2 model do not significantly enhance performance. However, a hybrid model that we propose combining these methods yields significantly better results, underscoring the merits of integrating classical and contemporary deep learning techniques in information retrieval in the domain of specialized scientific documents.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge