Robust Time-Series Retrieval Using Probabilistic Adaptive Segmental Alignment
Paper and Code
Sep 26, 2016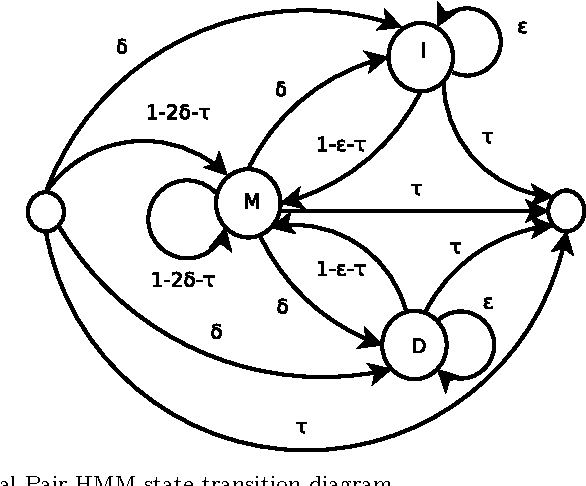

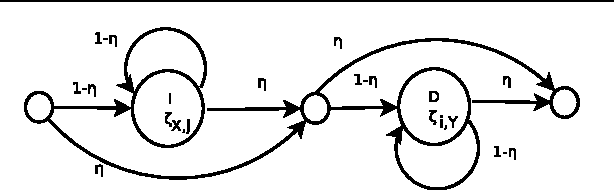
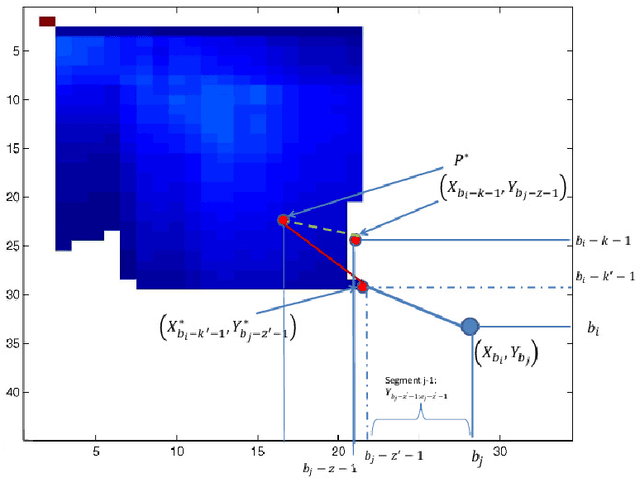
Traditional pairwise sequence alignment is based on matching individual samples from two sequences, under time monotonicity constraints. However, in many application settings matching subsequences (segments) instead of individual samples may bring in additional robustness to noise or local non-causal perturbations. This paper presents an approach to segmental sequence alignment that jointly segments and aligns two sequences, generalizing the traditional per-sample alignment. To accomplish this task, we introduce a distance metric between segments based on average pairwise distances and then present a modified pair-HMM (PHMM) that incorporates the proposed distance metric to solve the joint segmentation and alignment task. We also propose a relaxation to our model that improves the computational efficiency of the generic segmental PHMM. Our results demonstrate that this new measure of sequence similarity can lead to improved classification performance, while being resilient to noise, on a variety of sequence retrieval problems, from EEG to motion sequence classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge