Removal of Batch Effects using Generative Adversarial Networks
Paper and Code
Jan 20, 2019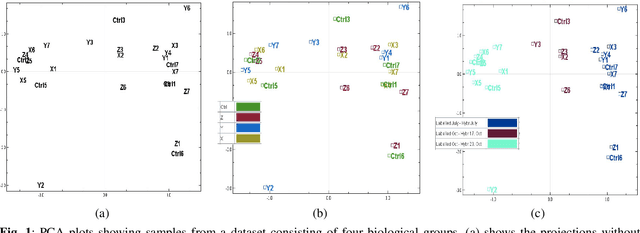
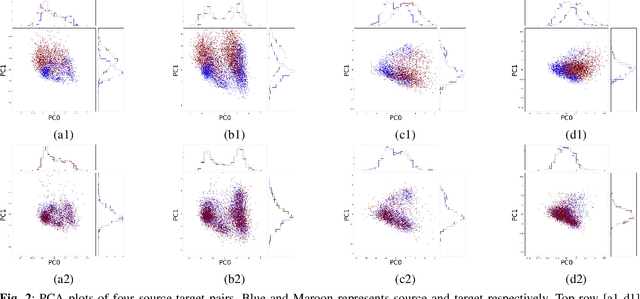
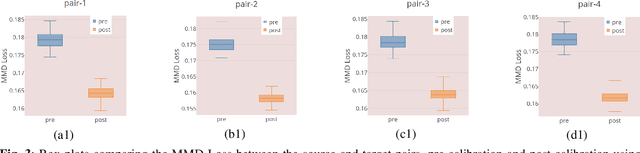
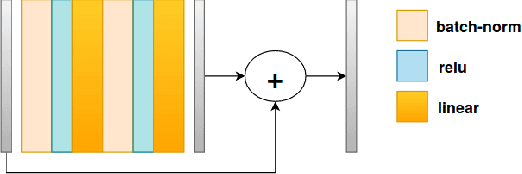
Many biological data analysis processes like Cytometry or Next Generation Sequencing (NGS) produce massive amounts of data which needs to be processed in batches for down-stream analysis. Such datasets are prone to technical variations due to difference in handling the batches possibly at different times, by different experimenters or under other different conditions. This adds variation to the batches coming from the same source sample. These variations are known as Batch Effects. It is possible that these variations and natural variations due to biology confound but such situations can be avoided by performing experiments in a carefully planned manner. Batch effects can hamper down-stream analysis and may also cause results to be inconclusive. Thus, it is essential to correct for these effects. Some recent methods propose deep learning based solution to solve this problem. We demonstrate that this can be solved using a novel Generative Adversarial Networks (GANs) based framework. The advantage of using this framework over other prior approaches is that here we do not require to choose a reproducing kernel and define its parameters.We demonstrate results of our framework on a Mass Cytometry dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge