Probing the Geometry of Data with Diffusion Fréchet Functions
Paper and Code
Mar 08, 2017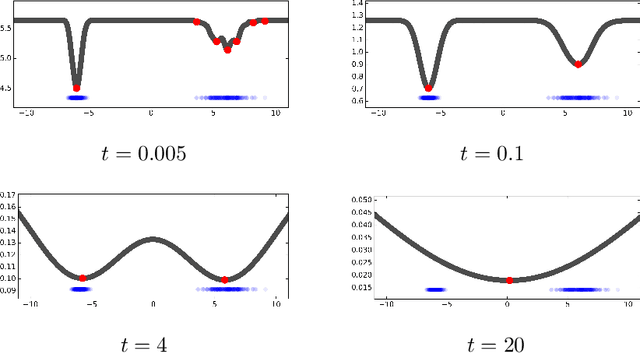
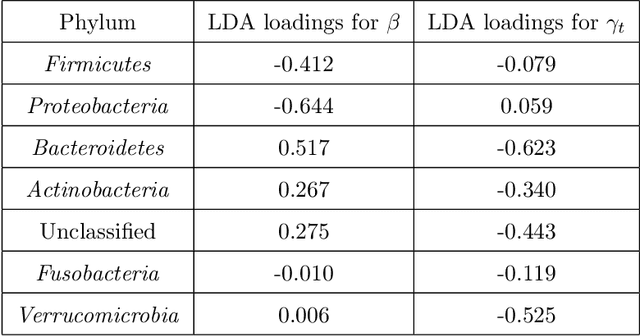
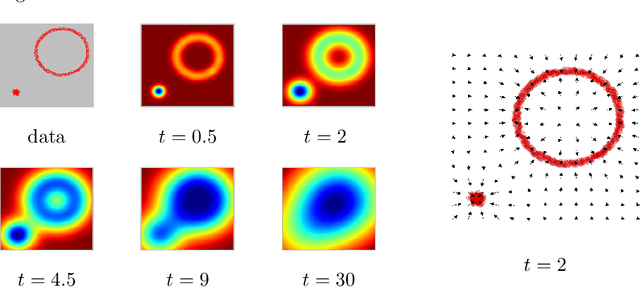
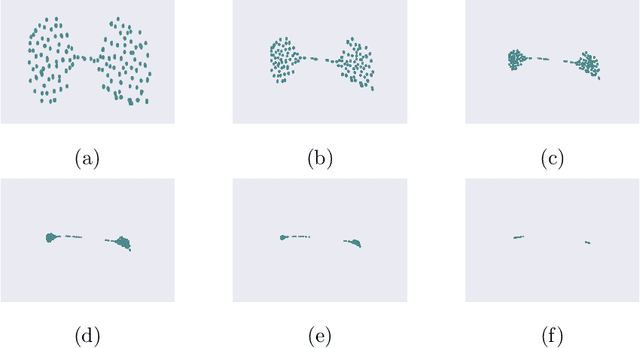
Many complex ecosystems, such as those formed by multiple microbial taxa, involve intricate interactions amongst various sub-communities. The most basic relationships are frequently modeled as co-occurrence networks in which the nodes represent the various players in the community and the weighted edges encode levels of interaction. In this setting, the composition of a community may be viewed as a probability distribution on the nodes of the network. This paper develops methods for modeling the organization of such data, as well as their Euclidean counterparts, across spatial scales. Using the notion of diffusion distance, we introduce diffusion Frechet functions and diffusion Frechet vectors associated with probability distributions on Euclidean space and the vertex set of a weighted network, respectively. We prove that these functional statistics are stable with respect to the Wasserstein distance between probability measures, thus yielding robust descriptors of their shapes. We apply the methodology to investigate bacterial communities in the human gut, seeking to characterize divergence from intestinal homeostasis in patients with Clostridium difficile infection (CDI) and the effects of fecal microbiota transplantation, a treatment used in CDI patients that has proven to be significantly more effective than traditional treatment with antibiotics. The proposed method proves useful in deriving a biomarker that might help elucidate the mechanisms that drive these processes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge